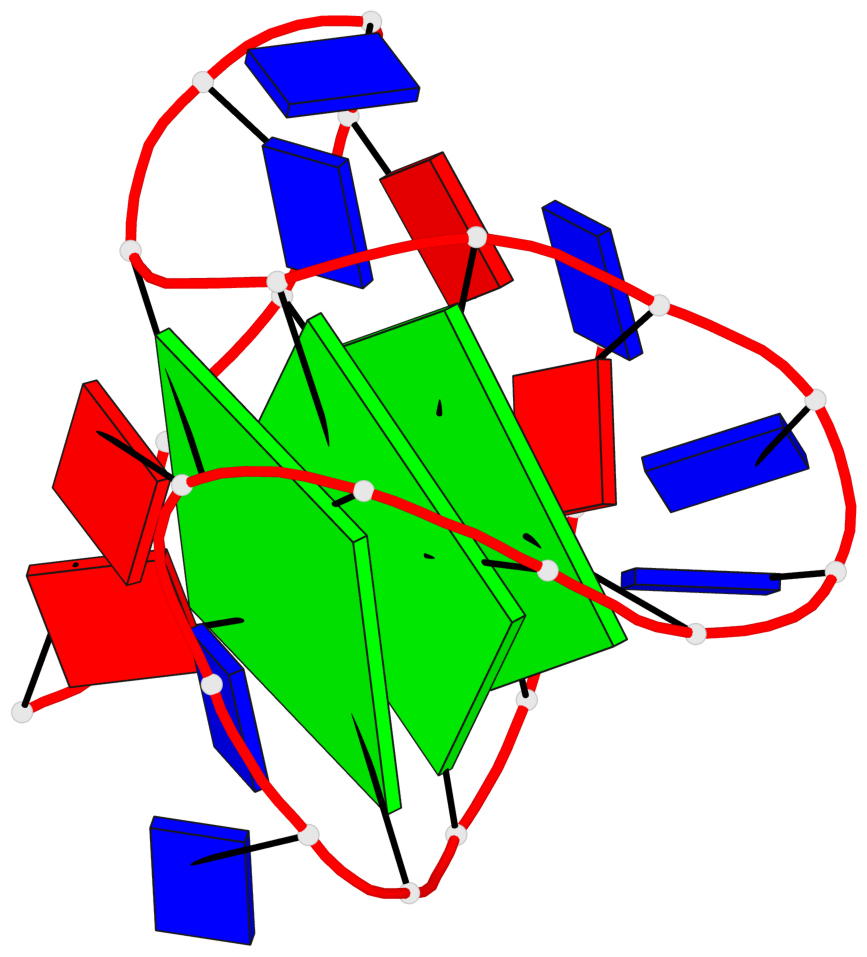
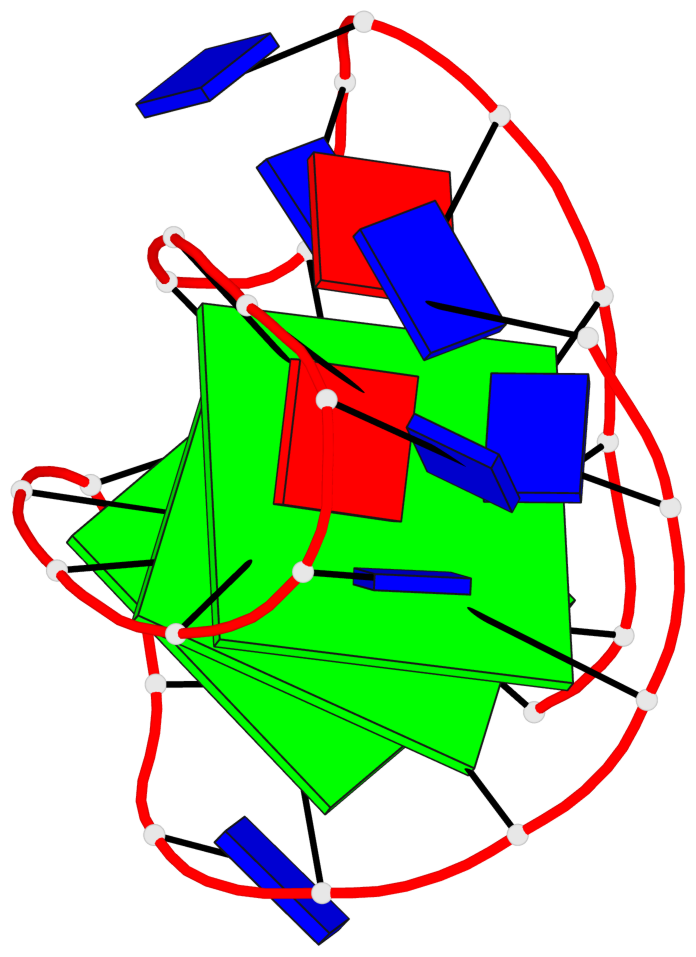
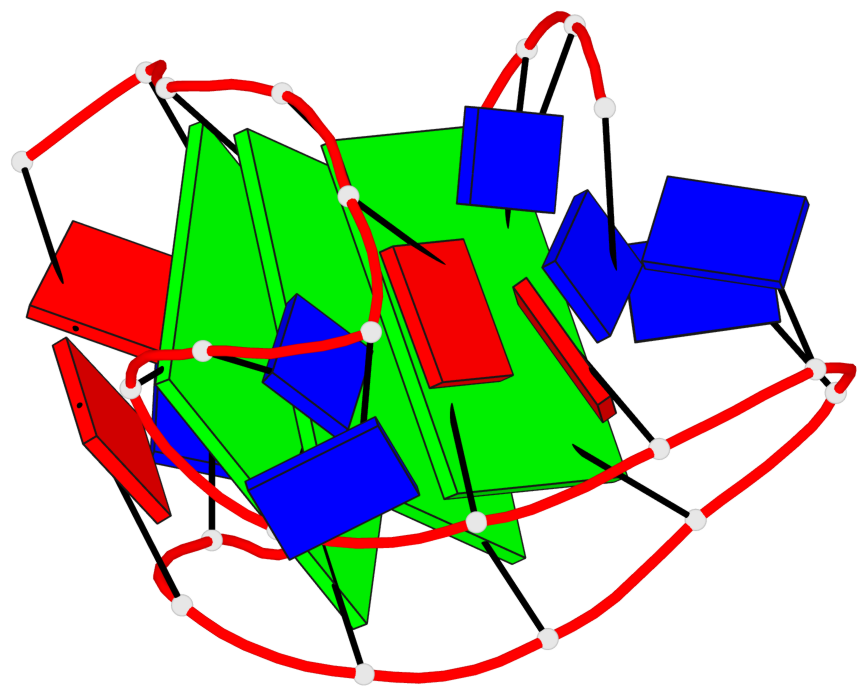
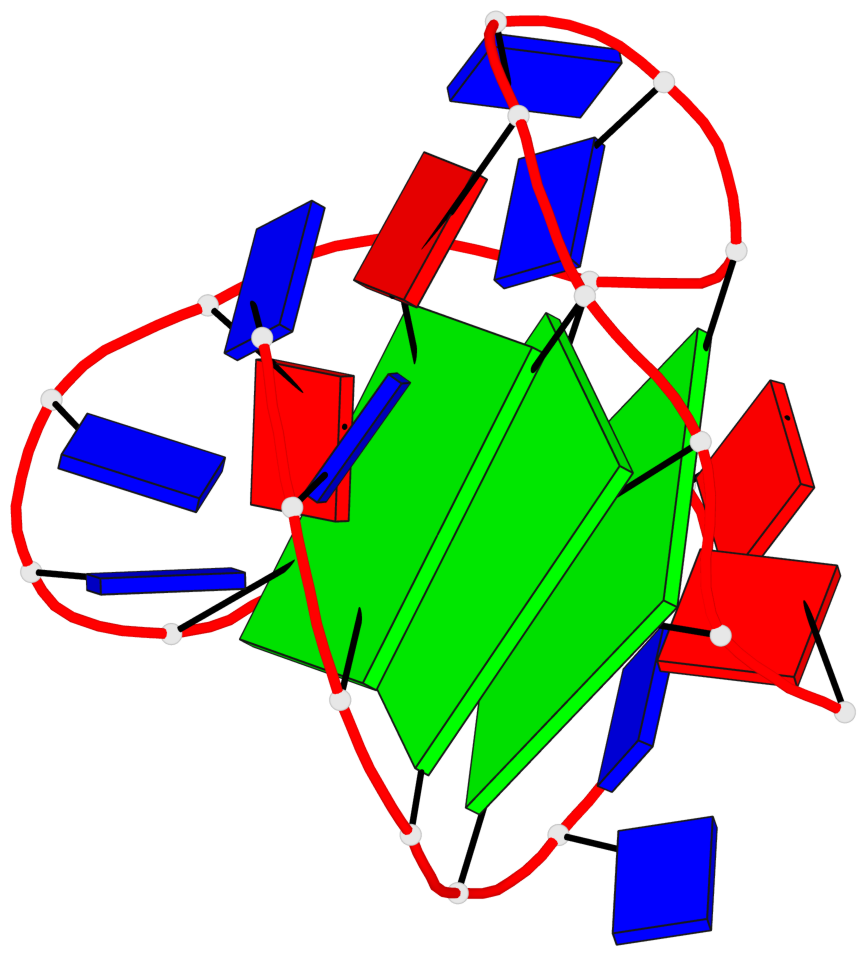
Detailed DSSR results for the G-quadruplex: PDB entry 6ia0
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 6ia0
- Class
- DNA
- Method
- NMR
- Summary
- Human telomeric g-quadruplex with 8-oxo-g substitution in the central g-quartet
- Reference
- Bielskute S, Plavec J, Podbevsek P (2019): "Impact of Oxidative Lesions on the Human Telomeric G-Quadruplex." J. Am. Chem. Soc., 141, 2594-2603. doi: 10.1021/jacs.8b12748.
- Abstract
- Telomere attrition is closely associated with cell aging and exposure to reactive oxygen species (ROS). While oxidation products of nucleotides have been studied extensively in the past, the underlying secondary/tertiary structural changes in DNA remain poorly understood. In this work, we systematically probed guanine positions in the human telomeric oligonucleotide sequence (hTel) by substitutions with the major product of ROS, 8-oxo-7,8-dihydroguanine (oxoG), and evaluated the G-quadruplex forming ability of such oligonucleotides. Due to reduced hydrogen-bonding capability caused by oxoG, a loss of G-quadruplex structure was observed for most oligonucleotides containing oxidative lesions. However, some positions in the hTel sequence were found to tolerate substitutions with oxoG. Due to oxo G's preference for the syn conformation, distinct responses were observed when replacing guanines with different glycosidic conformations. Accommodation of oxoG at sites originally in syn or anti in nonsubstituted hTel G-quadruplex requires a minor structural rearrangement or a major conformational shift, respectively. The system responds by retaining or switching to a fold where oxoG is in syn conformation. Most importantly, these G-quadruplex structures are still stable at physiological temperatures and should be considered detrimental in higher-order telomere structures.
- G4 notes
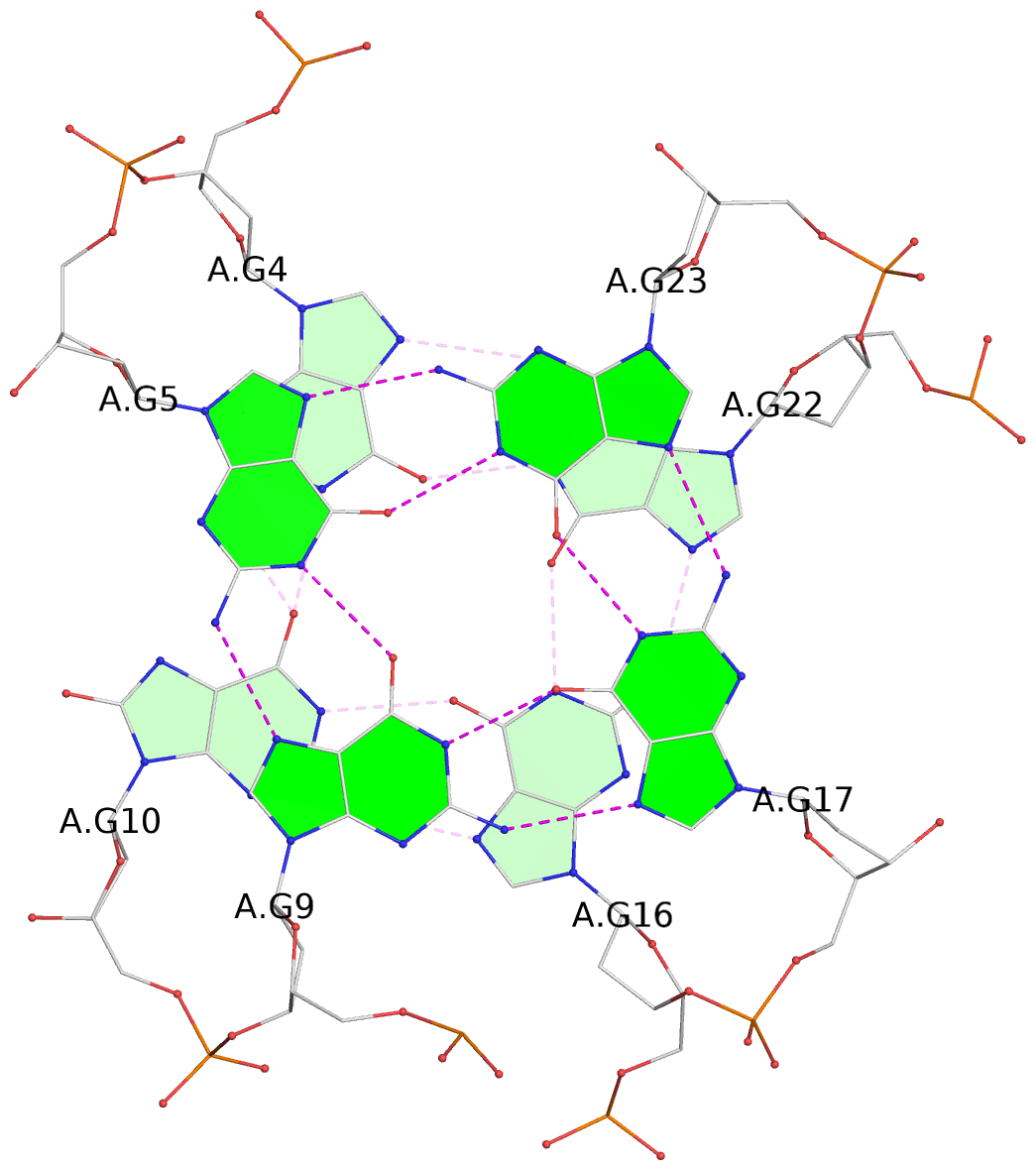
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-Lw-Ln-P), hybrid-2(3+1), UDUU
Base-block schematics in six views
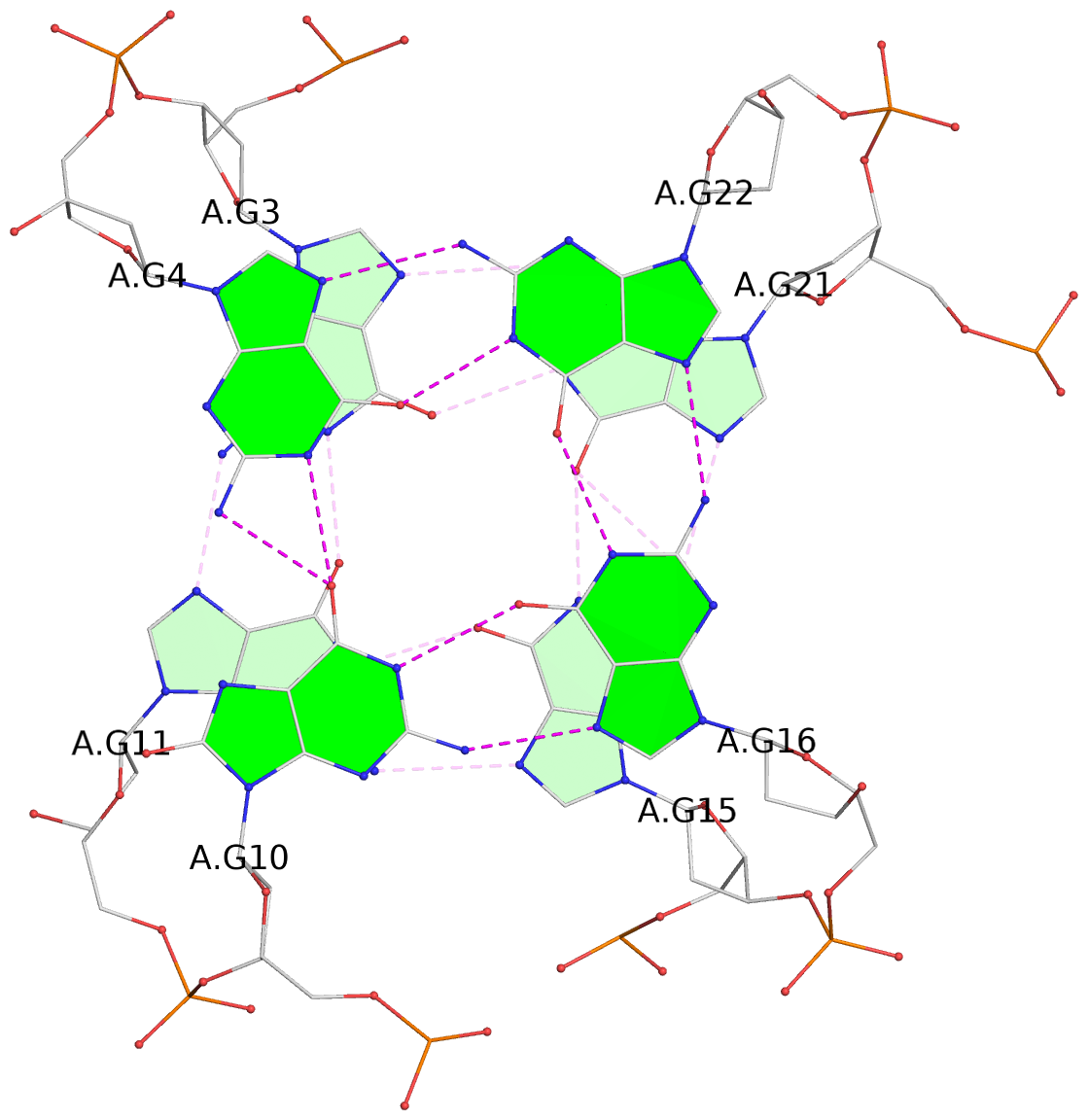
List of 3 G-tetrads
1 glyco-bond=-s-- sugar=---- groove=wn-- planarity=0.437 type=other nts=4 GGGG A.DG3,A.DG11,A.DG15,A.DG21 2 glyco-bond=-s-- sugar=---- groove=wn-- planarity=0.263 type=saddle nts=4 GgGG A.DG4,A.8OG10,A.DG16,A.DG22 3 glyco-bond=-s-- sugar=---- groove=wn-- planarity=0.233 type=other nts=4 GGGG A.DG5,A.DG9,A.DG17,A.DG23
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.