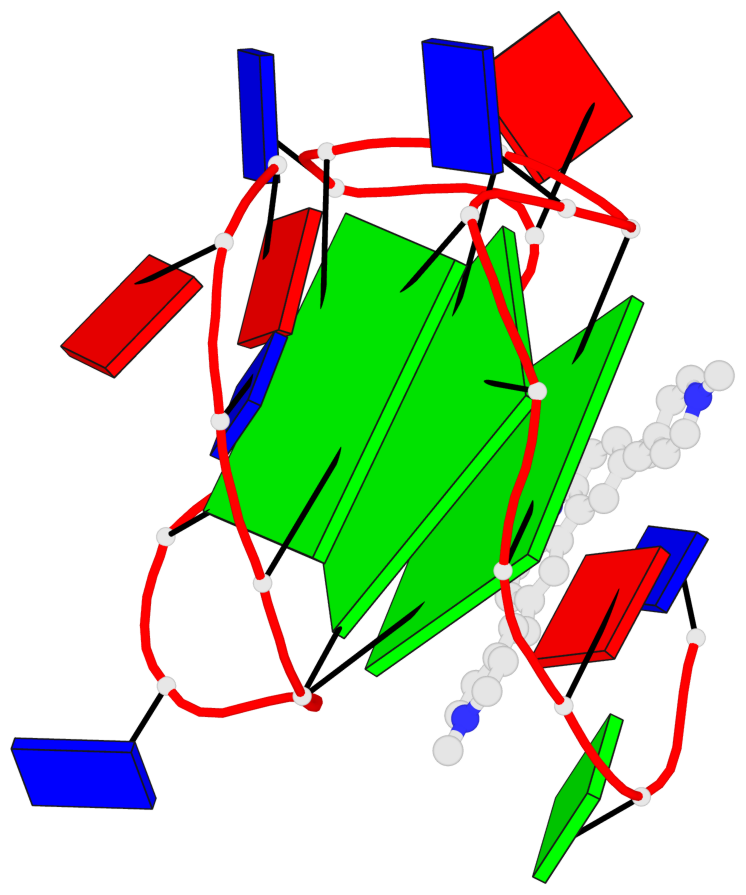
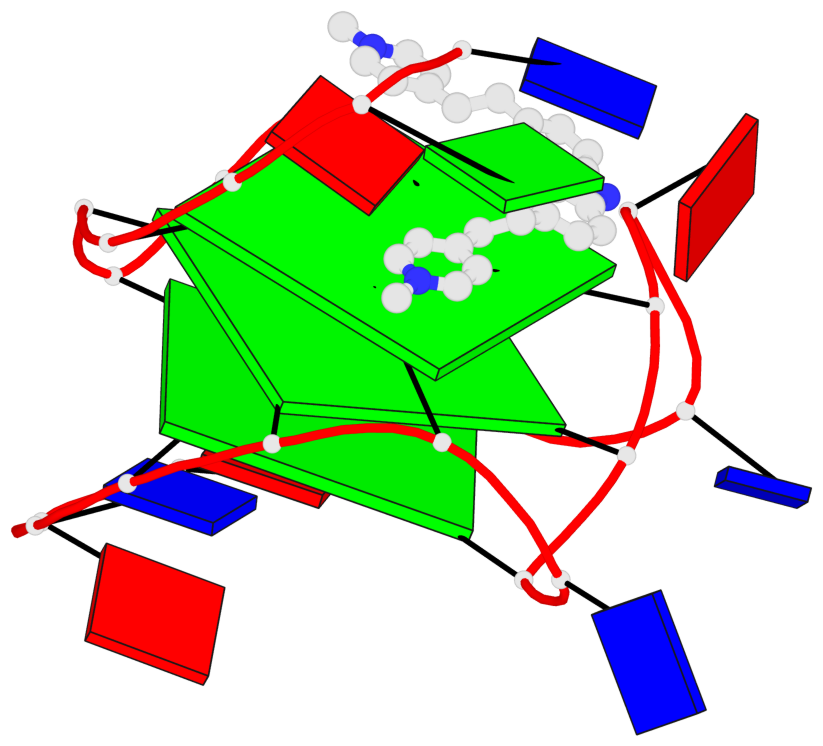
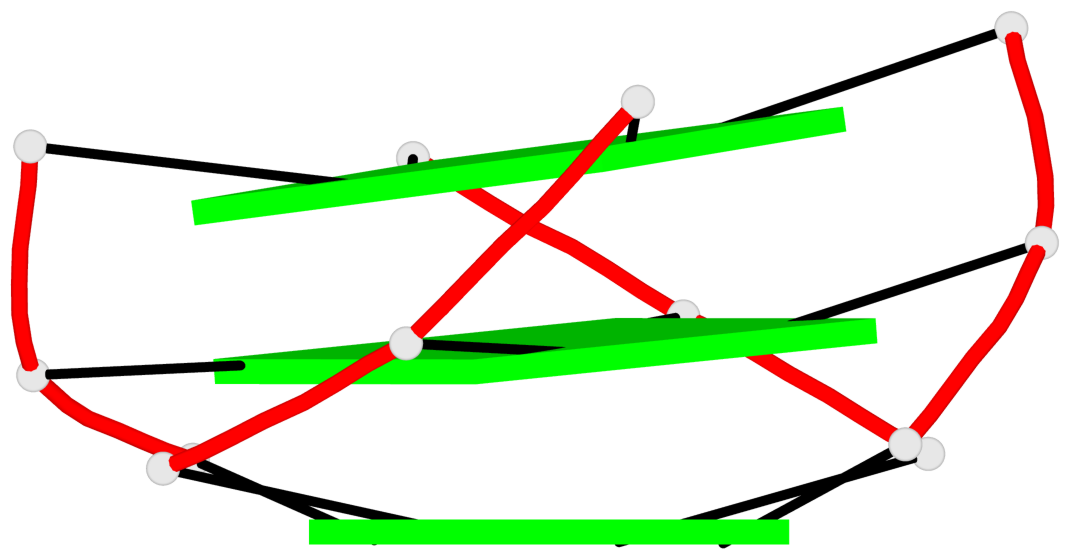
Detailed DSSR results for the G-quadruplex: PDB entry 6jj0
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 6jj0
- Class
- DNA
- Method
- NMR
- Summary
- NMR structure of the 1:1 complex of a carbazole derivative bmvc bound to c-myc g-quadruplex
- Reference
- Liu W, Lin C, Wu G, Dai J, Chang TC, Yang D (2019): "Structures of 1:1 and 2:1 complexes of BMVC and MYC promoter G-quadruplex reveal a mechanism of ligand conformation adjustment for G4-recognition." Nucleic Acids Res., 47, 11931-11942. doi: 10.1093/nar/gkz1015.
- Abstract
- BMVC is the first fluorescent probe designed to detect G-quadruplexes (G4s) in vivo. The MYC oncogene promoter forms a G4 (MycG4) which acts as a transcription silencer. Here, we report the high-affinity and specific binding of BMVC to MycG4 with unusual slow-exchange rates on the NMR timescale. We also show that BMVC represses MYC in cancer cells. We determined the solution structures of the 1:1 and 2:1 BMVC-MycG4 complexes. BMVC first binds the 5'-end of MycG4 to form a 1:1 complex with a well-defined structure. At higher ratio, BMVC also binds the 3'-end to form a second complex. In both complexes, the crescent-shaped BMVC recruits a flanking DNA residue to form a BMVC-base plane stacking over the external G-tetrad. Remarkably, BMVC adjusts its conformation to a contracted form to match the G-tetrad for an optimal stacking interaction. This is the first structural example showing the importance of ligand conformational adjustment in G4 recognition. BMVC binds the more accessible 5'-end with higher affinity, whereas sequence specificity is present at the weaker-binding 3'-site. Our structures provide insights into specific recognition of MycG4 by BMVC and useful information for design of G4-targeted anticancer drugs and fluorescent probes.
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-P-P-P), parallel(4+0), UUUU
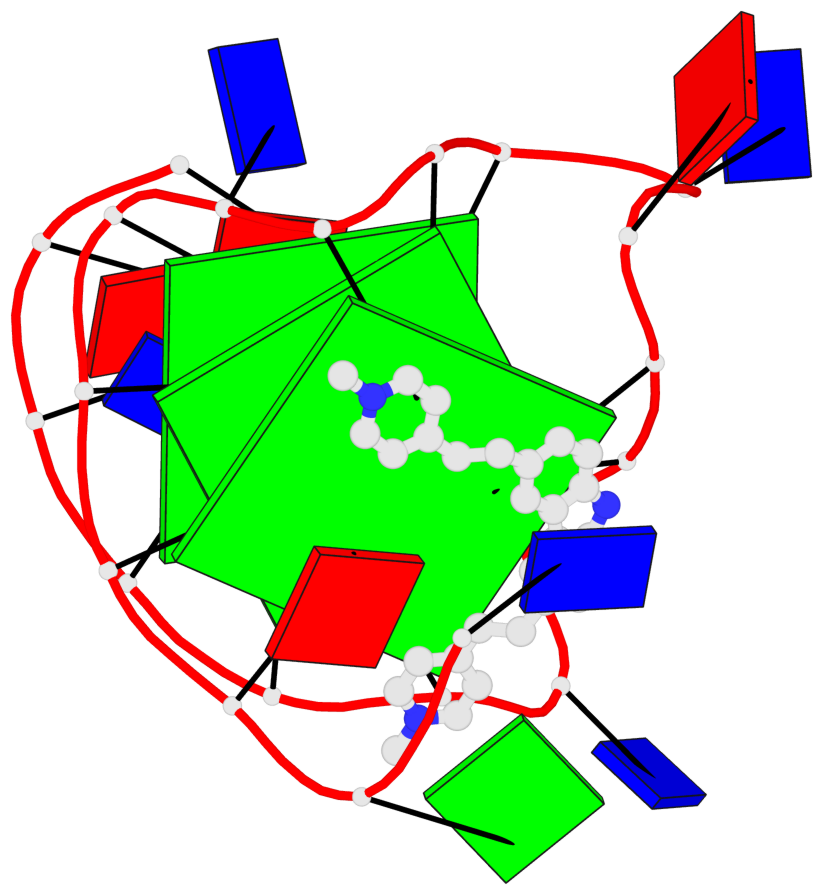
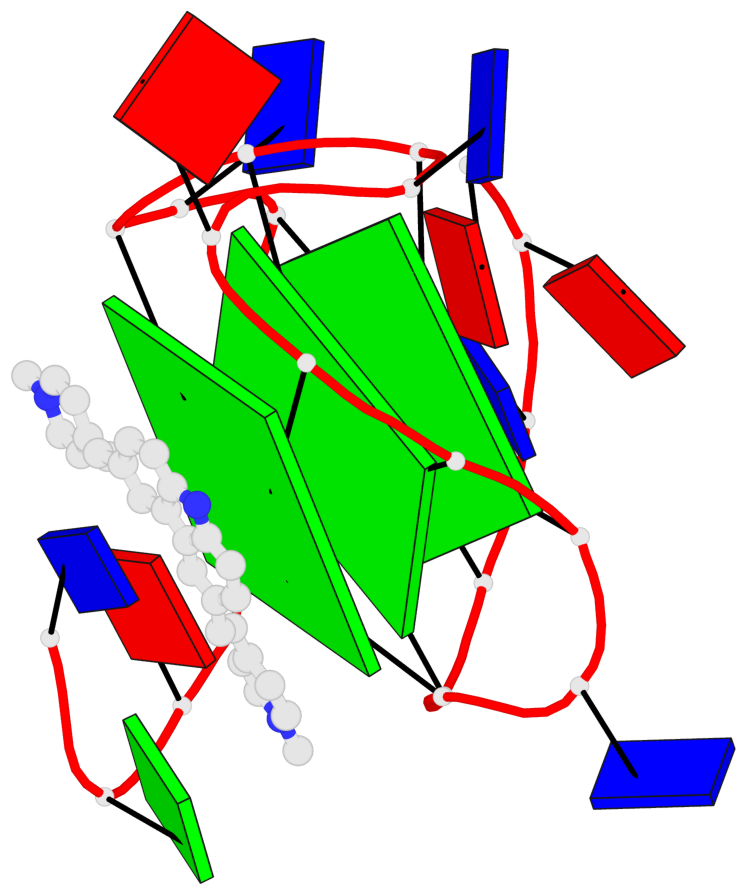
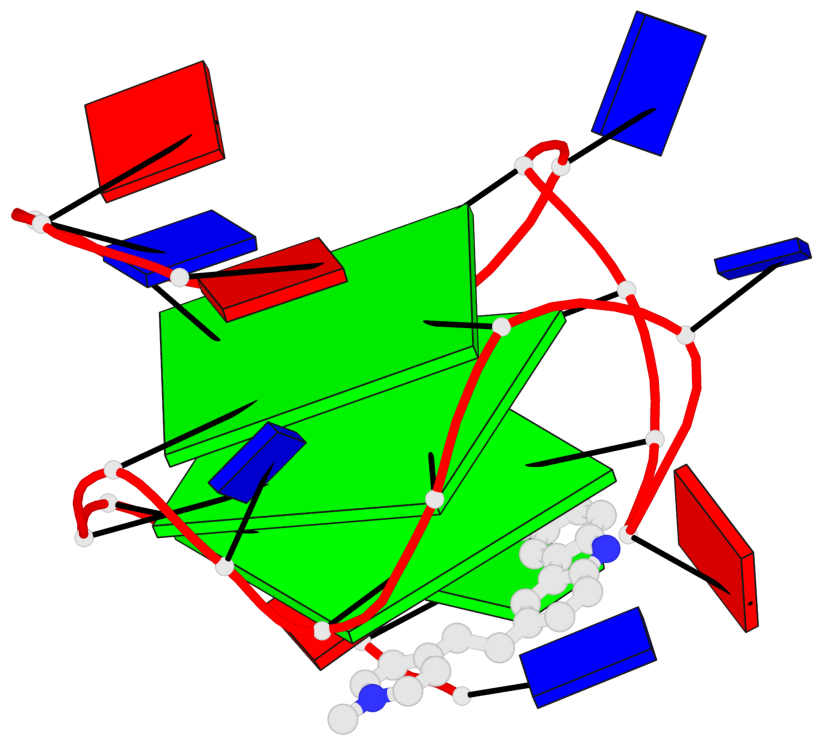
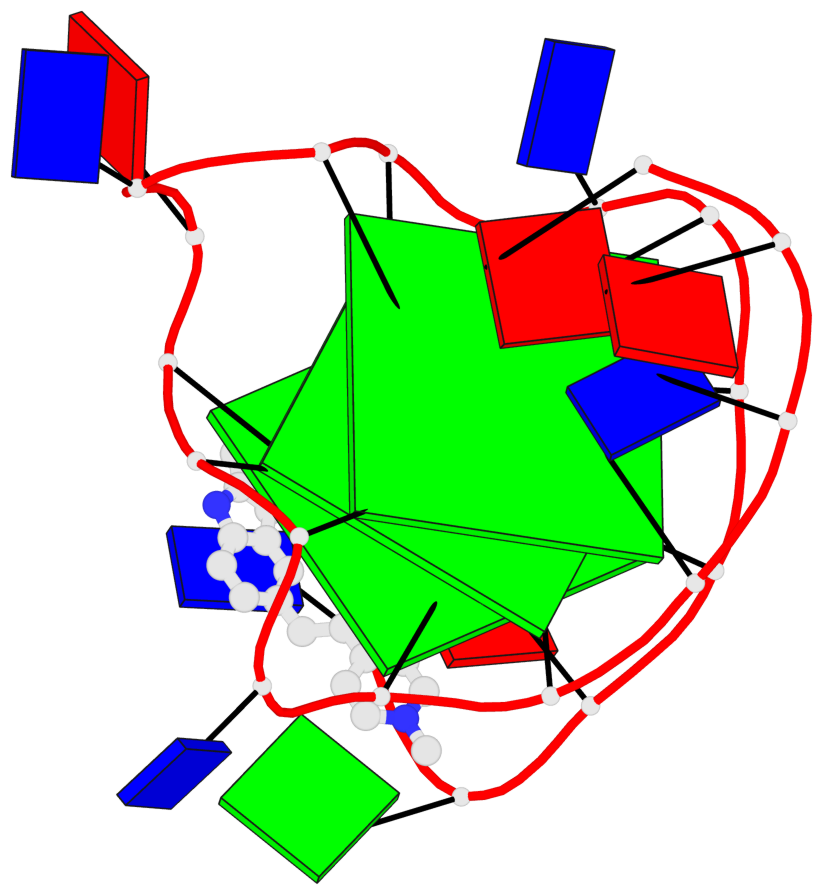
Base-block schematics in six views
List of 3 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.218 type=other nts=4 GGGG A.DG7,A.DG11,A.DG16,A.DG20 2 glyco-bond=---- sugar=---- groove=---- planarity=0.173 type=other nts=4 GGGG A.DG8,A.DG12,A.DG17,A.DG21 3 glyco-bond=---- sugar=---3 groove=---- planarity=0.056 type=planar nts=4 GGGG A.DG9,A.DG13,A.DG18,A.DG22
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.