Detailed DSSR results for the G-quadruplex: PDB entry 6jkn
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 6jkn
- Class
- DNA
- Method
- X-ray (1.401 Å)
- Summary
- Crystal structure of g-quadruplex formed by bromo-substituted human telomeric DNA
- Reference
- Geng Y, Liu C, Zhou B, Cai Q, Miao H, Shi X, Xu N, You Y, Fung CP, Din RU, Zhu G (2019): "The crystal structure of an antiparallel chair-type G-quadruplex formed by Bromo-substituted human telomeric DNA." Nucleic Acids Res., 47, 5395-5404. doi: 10.1093/nar/gkz221.
- Abstract
- Human telomeric guanine-rich DNA, which could adopt different G-quadruplex structures, plays important roles in protecting the cell from recombination and degradation. Although many of these structures were determined, the chair-type G-quadruplex structure remains elusive. Here, we present a crystal structure of the G-quadruplex composed of the human telomeric sequence d[GGGTTAGG8GTTAGGGTTAGG20G] with two dG to 8Br-dG substitutions at positions 8 and 20 with syn conformation in the K+ solution. It forms a novel three-layer chair-type G-quadruplex with two linking trinucleotide loops. Particularly, T5 and T17 are coplanar with two water molecules stacking on the G-tetrad layer in a sandwich-like mode through a coordinating K+ ion and an A6•A18 base pair. While a twisted Hoogsteen A12•T10 base pair caps on the top of G-tetrad core. The three linking TTA loops are edgewise and each DNA strand has two antiparallel adjacent strands. Our findings contribute to a deeper understanding and highlight the unique roles of loop and water molecule in the folding of the G-quadruplex.
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-Lw-Ln-Lw), chair(2+2), UDUD
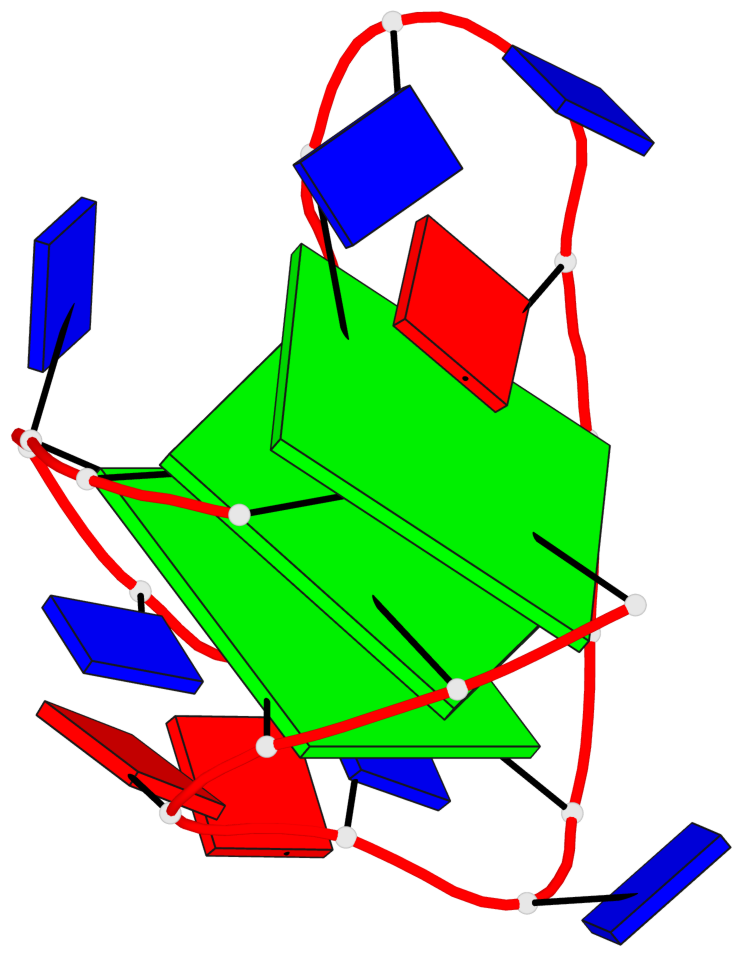
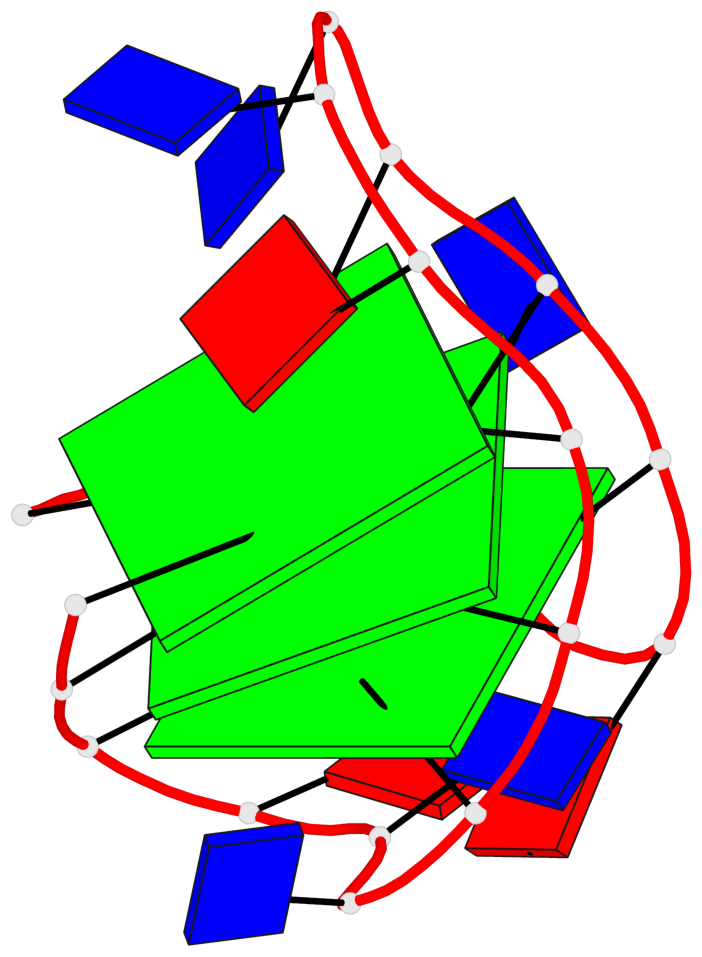
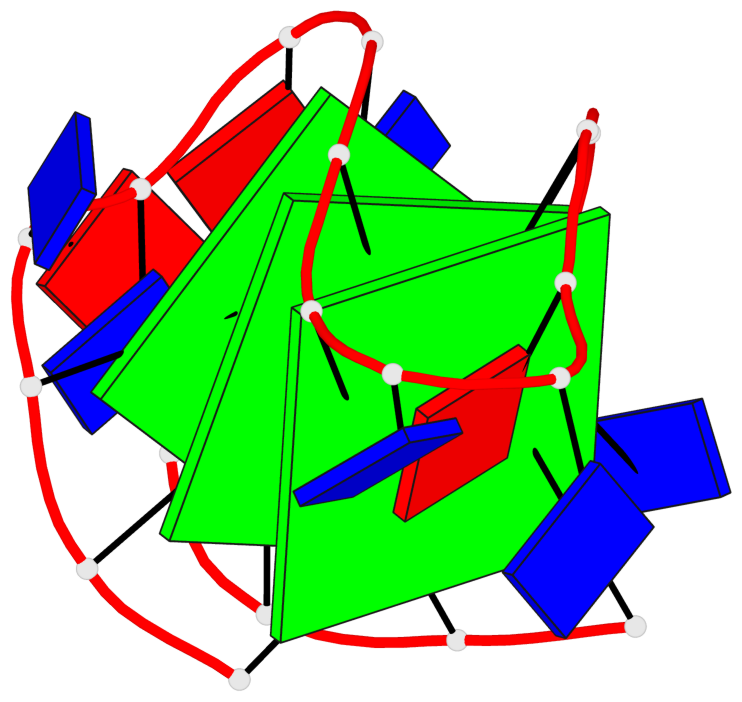
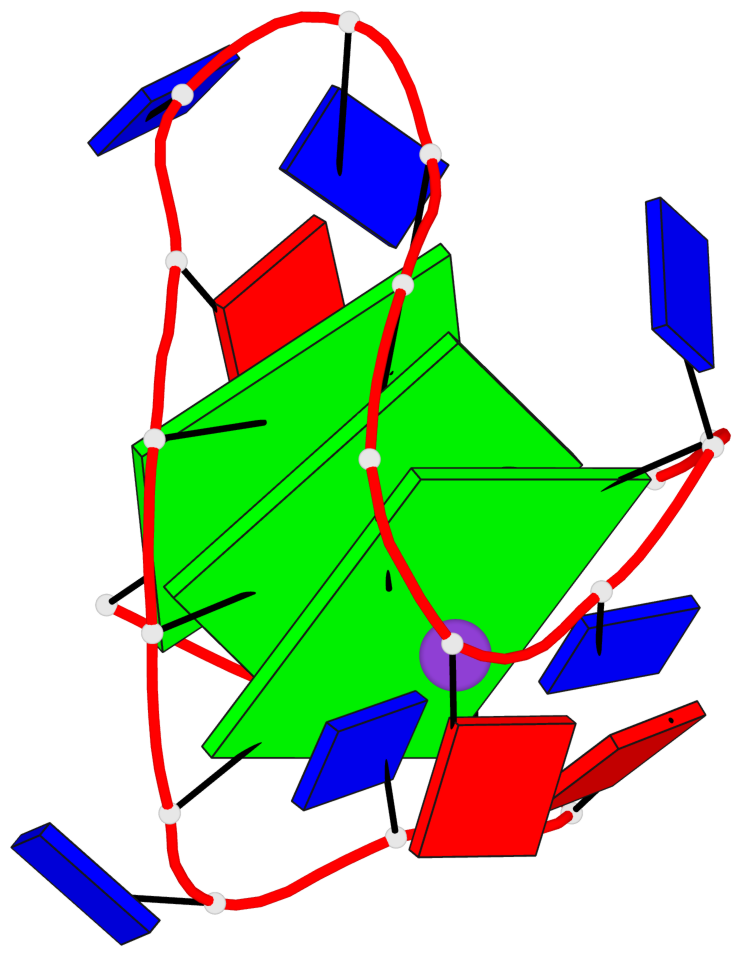
Base-block schematics in six views
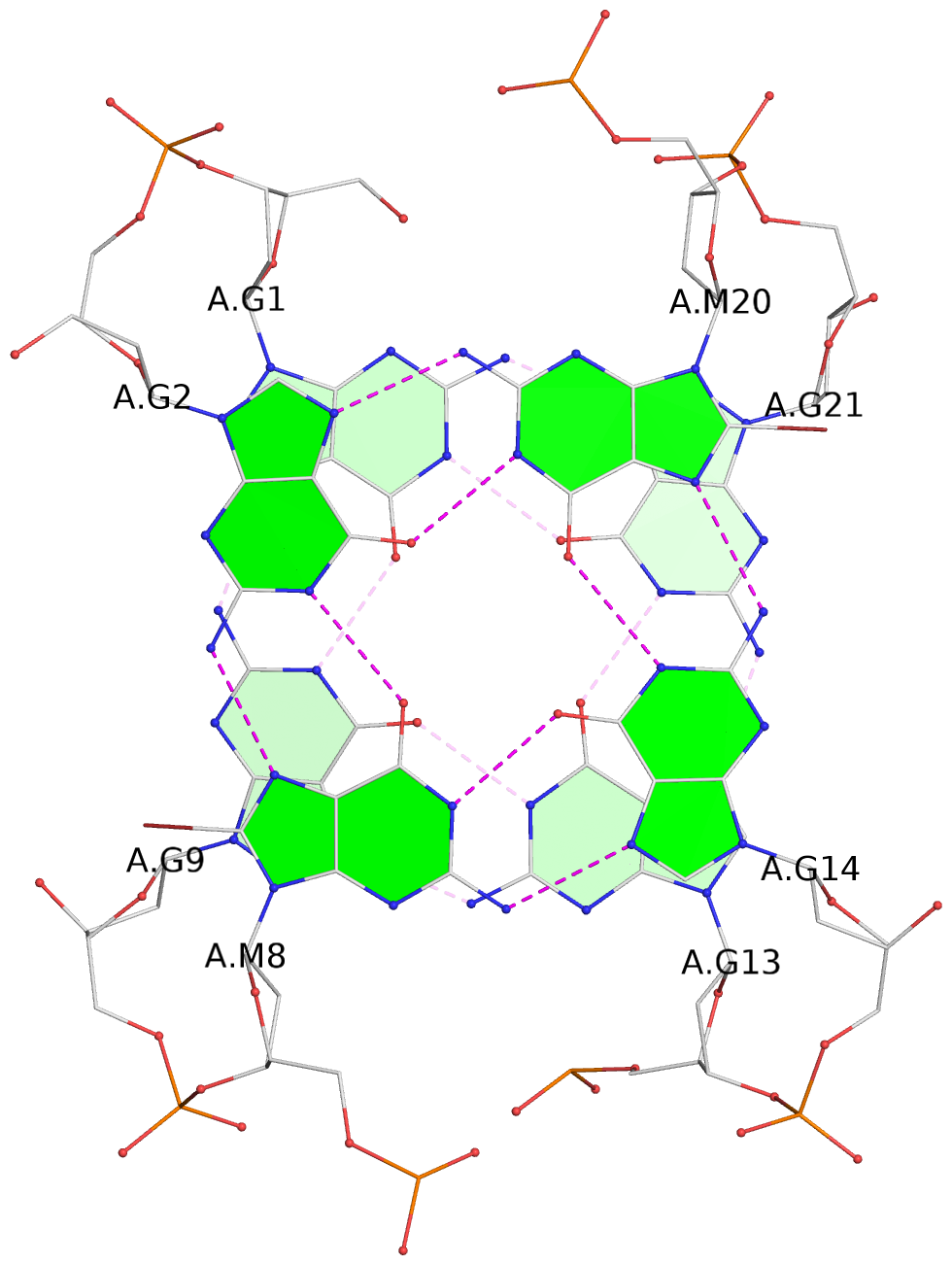
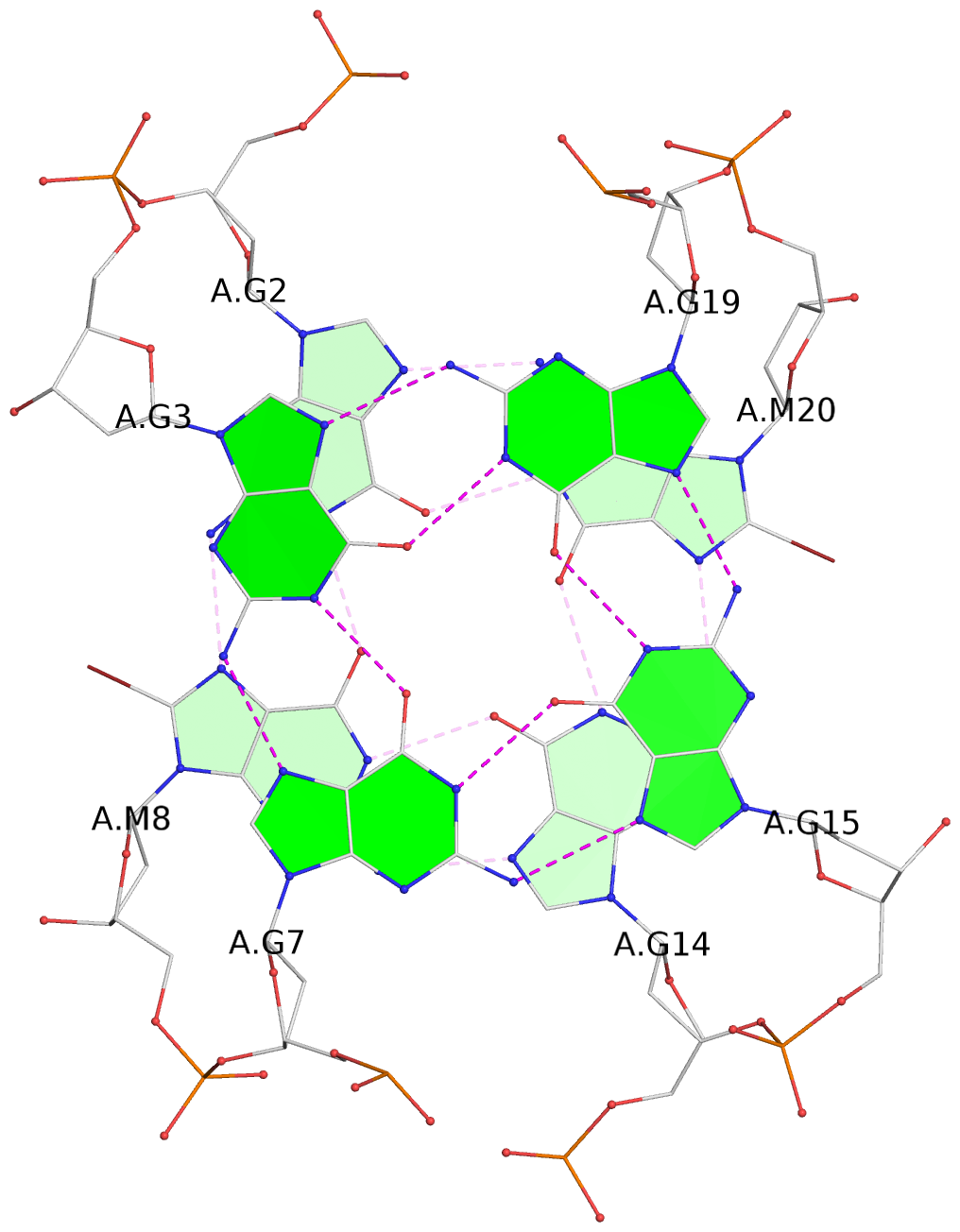
List of 3 G-tetrads
1 glyco-bond=s-s- sugar=---3 groove=wnwn planarity=0.607 type=bowl-2 nts=4 GGGG A.DG1,A.DG9,A.DG13,A.DG21 2 glyco-bond=-s-s sugar=---. groove=wnwn planarity=0.531 type=saddle nts=4 GgGg A.DG2,A.BGM8,A.DG14,A.BGM20 3 glyco-bond=-s-s sugar=---- groove=wnwn planarity=0.341 type=other nts=4 GGGG A.DG3,A.DG7,A.DG15,A.DG19
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.