Detailed DSSR results for the G-quadruplex: PDB entry 6jwe
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 6jwe
- Class
- DNA
- Method
- NMR
- Summary
- Structure of ret g-quadruplex in complex with colchicine
- Reference
- Wang F, Wang C, Liu Y, Lan W, Han H, Wang R, Huang S, Cao C (2020): "Colchicine selective interaction with oncogene RET G-quadruplex revealed by NMR." Chem.Commun.(Camb.), 56, 2099-2102. doi: 10.1039/d0cc00221f.
- Abstract
- G-quadruplexes (G4s) are frequently formed in the promoter regions of oncogenes, considered as promising drug targets for anticancer therapy. Due to high structure similarity of G4s, discovering ligands selectively interacting with only one G4 is extremely difficult. Here, mainly by NMR, we report that colchicine selectively binds to oncogene RET G4-DNA.
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-P-P-P), parallel(4+0), UUUU
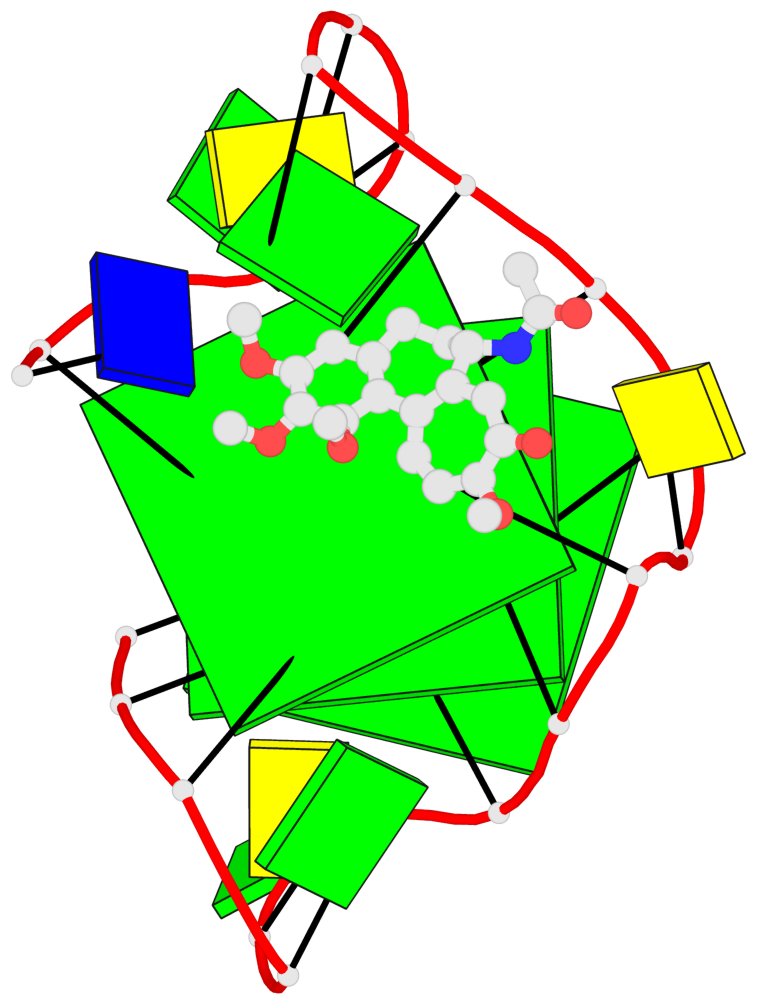
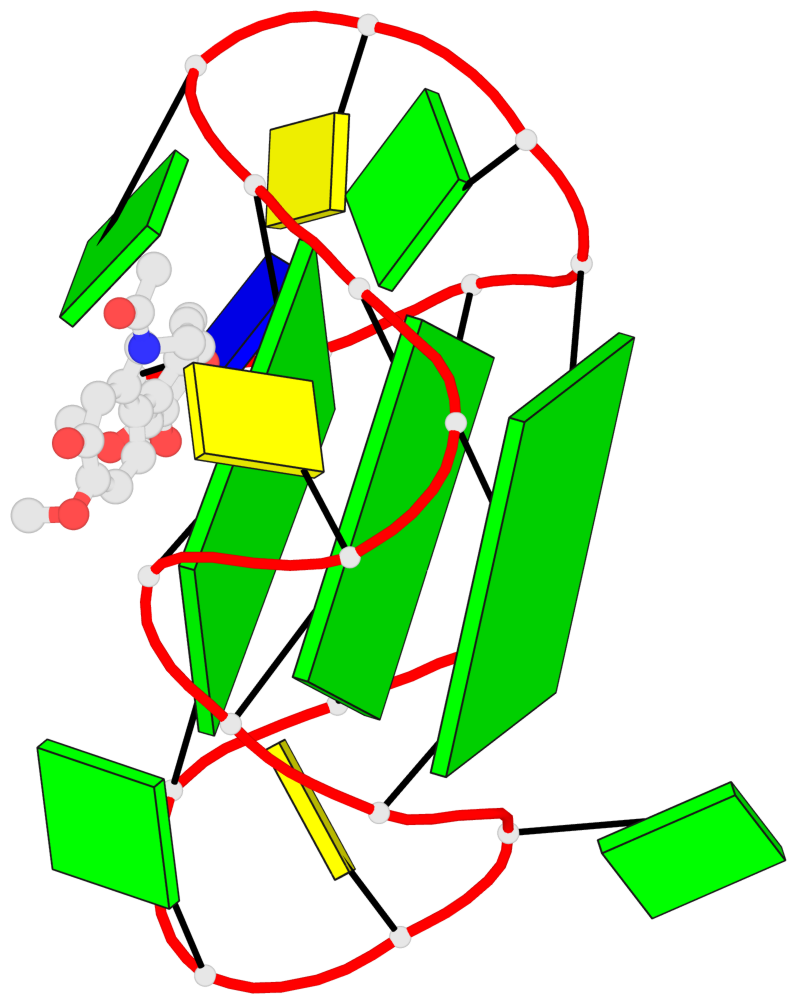
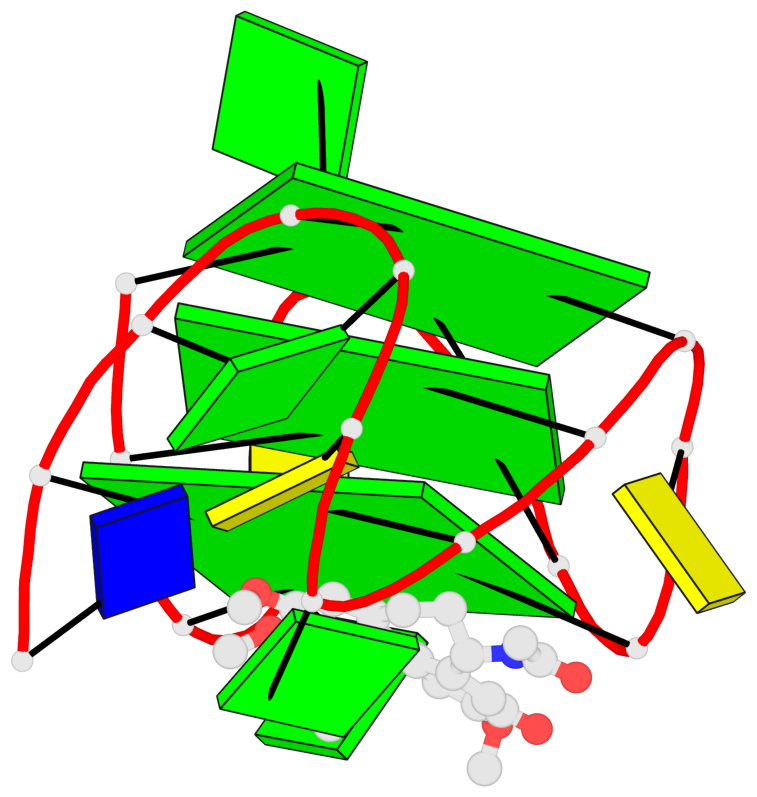
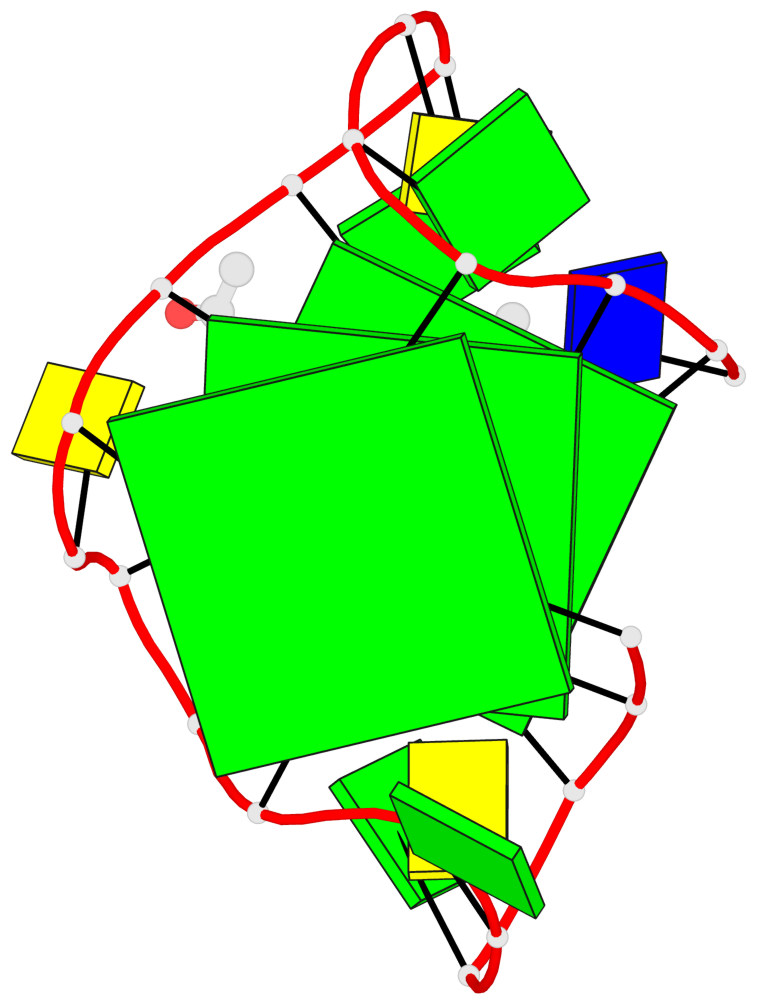
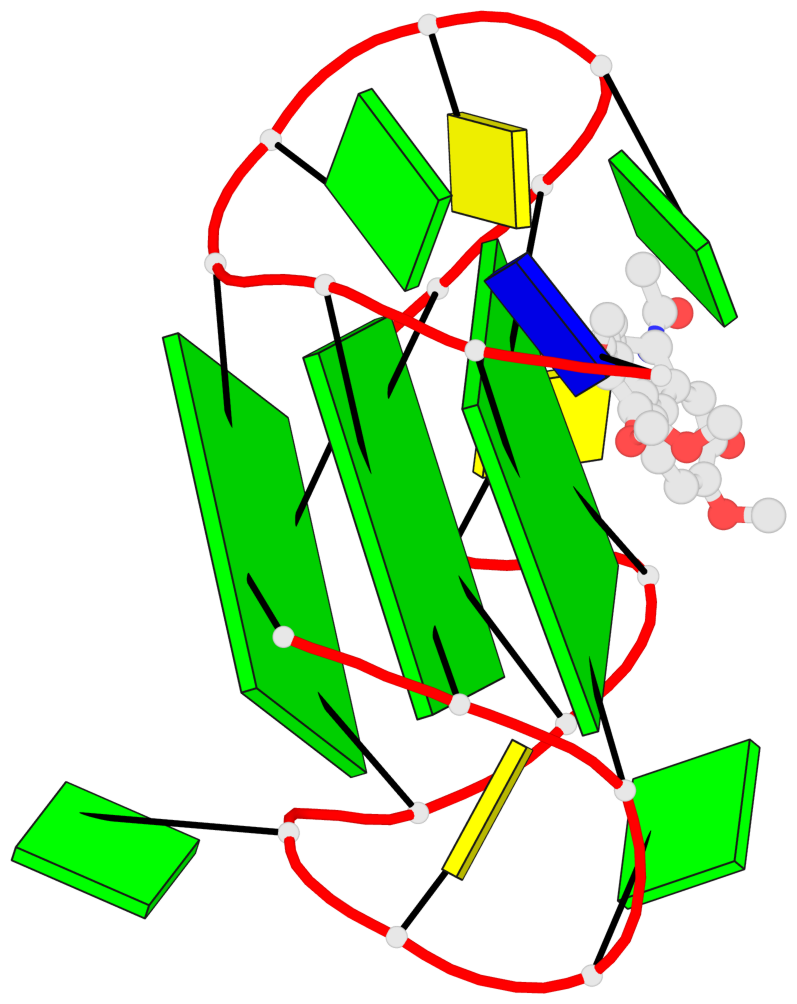
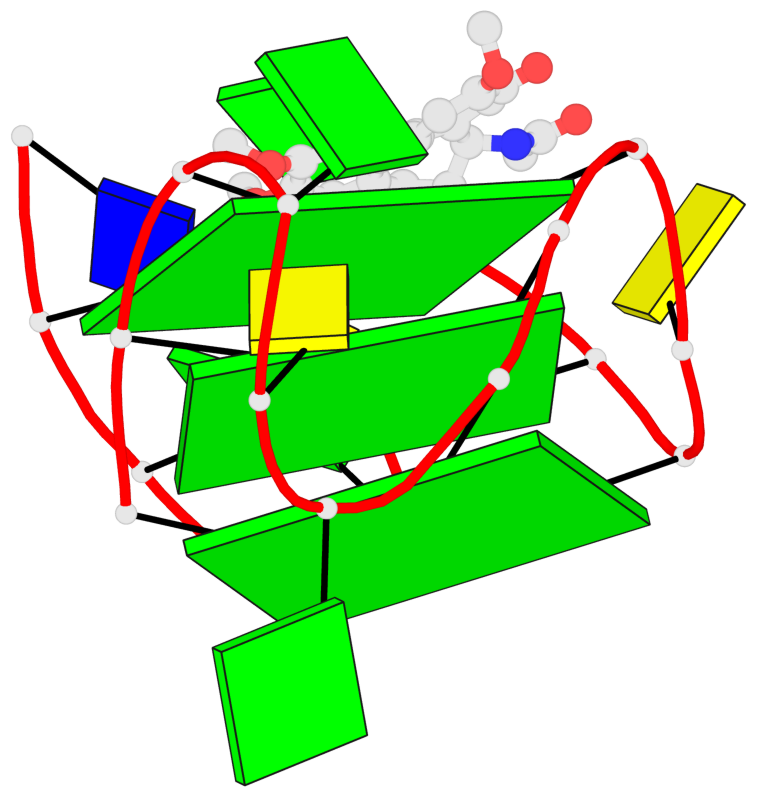
Base-block schematics in six views
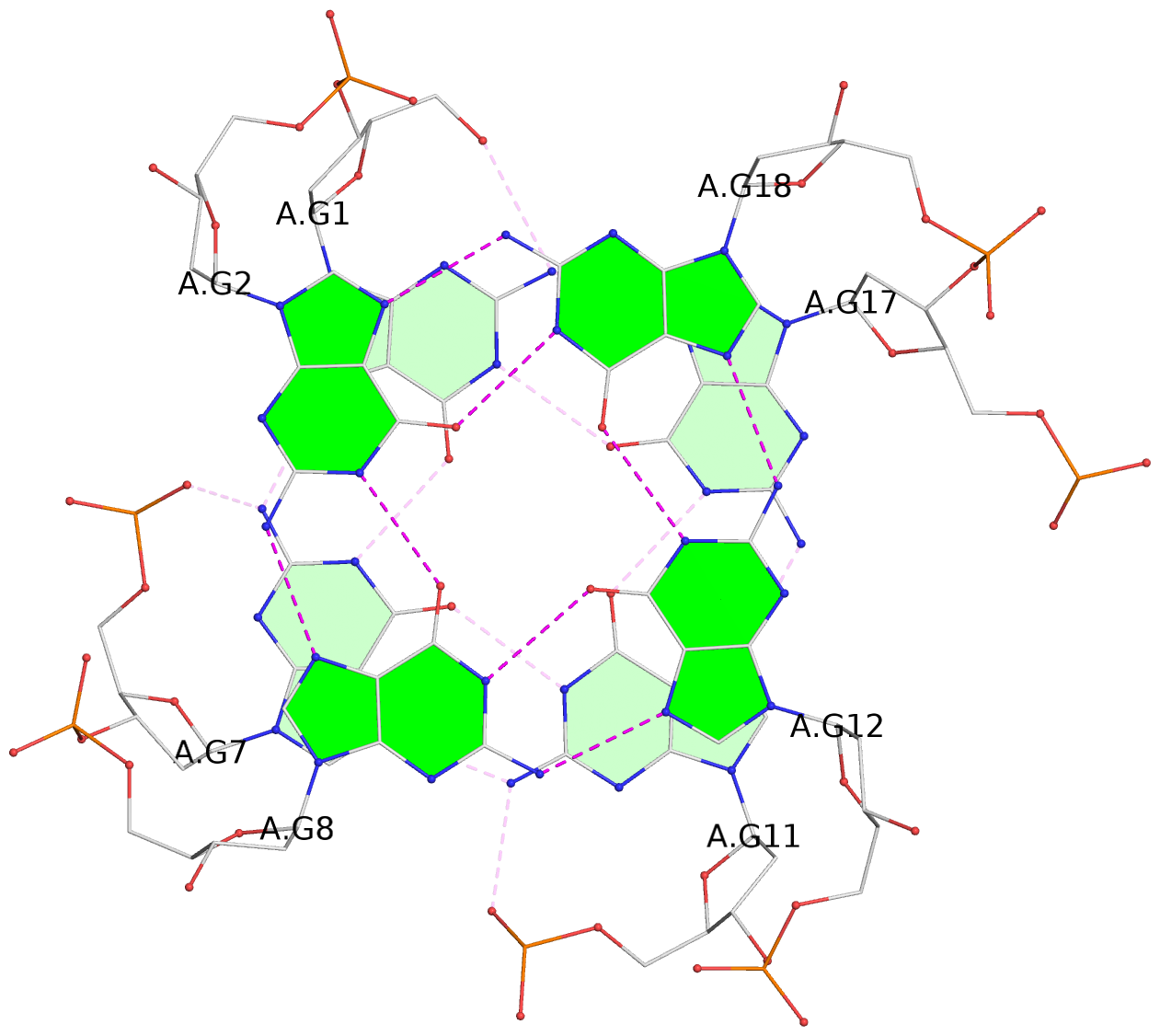
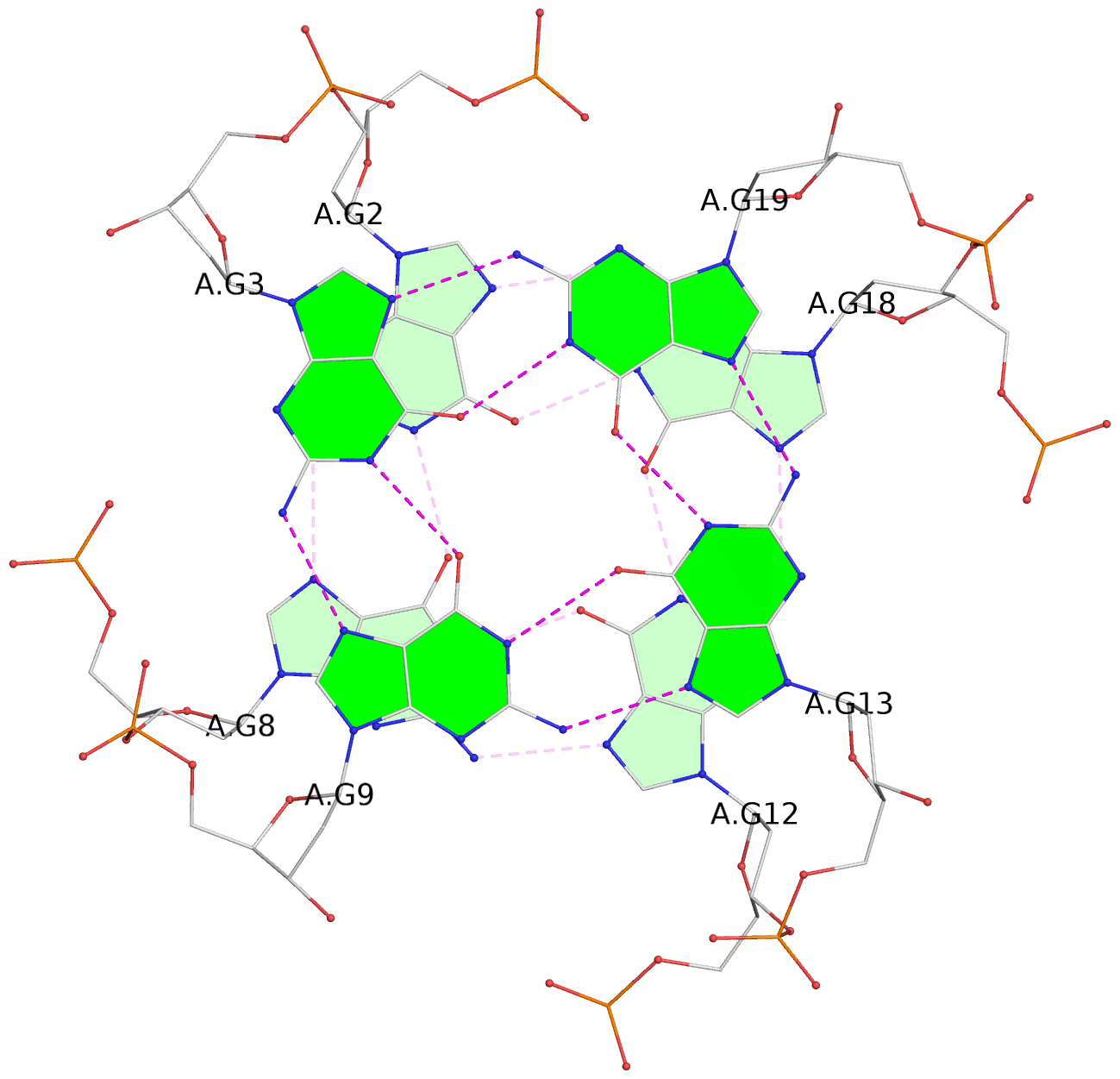
List of 3 G-tetrads
1 glyco-bond=ssss sugar=.33. groove=---- planarity=0.086 type=planar nts=4 GGGG A.DG1,A.DG7,A.DG11,A.DG17 2 glyco-bond=---- sugar=3333 groove=---- planarity=0.063 type=planar nts=4 GGGG A.DG2,A.DG8,A.DG12,A.DG18 3 glyco-bond=---- sugar=--33 groove=---- planarity=0.122 type=planar nts=4 GGGG A.DG3,A.DG9,A.DG13,A.DG19
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.