Detailed DSSR results for the G-quadruplex: PDB entry 6k84
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 6k84
- Class
- RNA
- Method
- NMR
- Summary
- Structure of anti-prion RNA aptamer
- Reference
- Mashima T, Lee JH, Kamatari YO, Hayashi T, Nagata T, Nishikawa F, Nishikawa S, Kinoshita M, Kuwata K, Katahira M (2020): "Development and structural determination of an anti-PrPCaptamer that blocks pathological conformational conversion of prion protein." Sci Rep, 10, 4934. doi: 10.1038/s41598-020-61966-4.
- Abstract
- Prion diseases comprise a fatal neuropathy caused by the conversion of prion protein from a cellular (PrPC) to a pathological (PrPSc) isoform. Previously, we obtained an RNA aptamer, r(GGAGGAGGAGGA) (R12), that folds into a unique G-quadruplex. The R12 homodimer binds to a PrPC molecule, inhibiting PrPC-to-PrPSc conversion. Here, we developed a new RNA aptamer, r(GGAGGAGGAGGAGGAGGAGGAGGA) (R24), where two R12s are tandemly connected. The 50% inhibitory concentration for the formation of PrPSc (IC50) of R24 in scrapie-infected cell lines was ca. 100 nM, i.e., much lower than that of R12 by two orders. Except for some antibodies, R24 exhibited the lowest recorded IC50 and the highest anti-prion activity. We also developed a related aptamer, r(GGAGGAGGAGGA-A-GGAGGAGGAGGA) (R12-A-R12), IC50 being ca. 500 nM. The structure of a single R12-A-R12 molecule determined by NMR resembled that of the R12 homodimer. The quadruplex structure of either R24 or R12-A-R12 is unimolecular, and therefore the structure could be stably formed when they are administered to a prion-infected cell culture. This may be the reason they can exert high anti-prion activity.
- G4 notes
- 4 G-tetrads, 1 G4 helix, 2 G4 stems, 1 G4 coaxial stack, 2(-P-P-P), parallel(4+0), UUUU, coaxial interfaces: 5'/5'
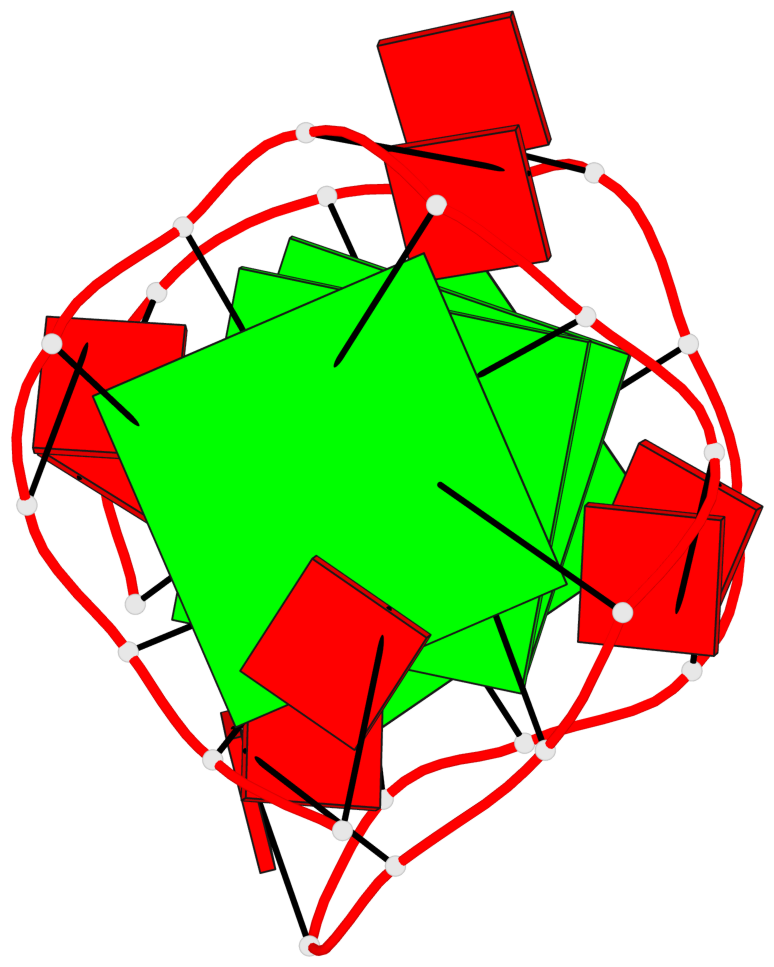
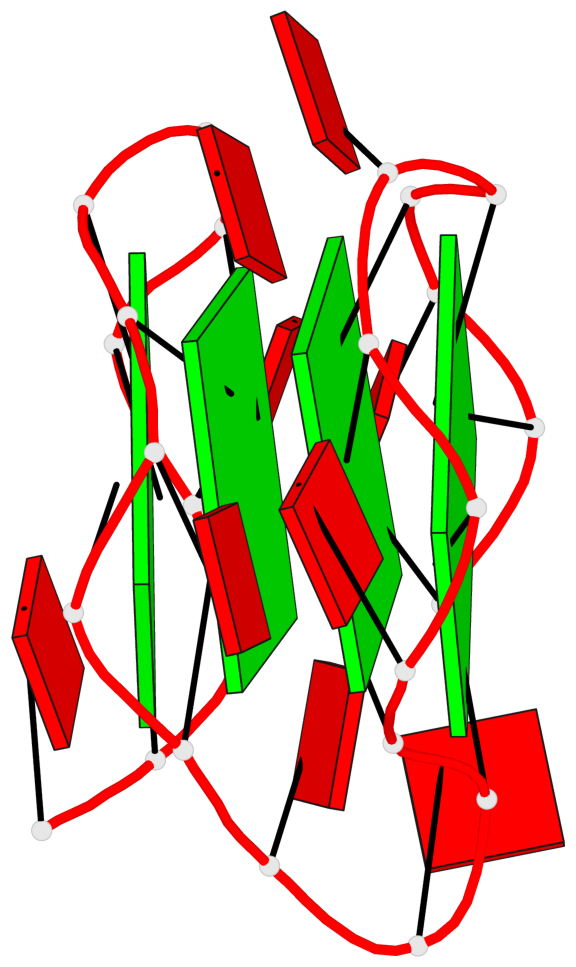
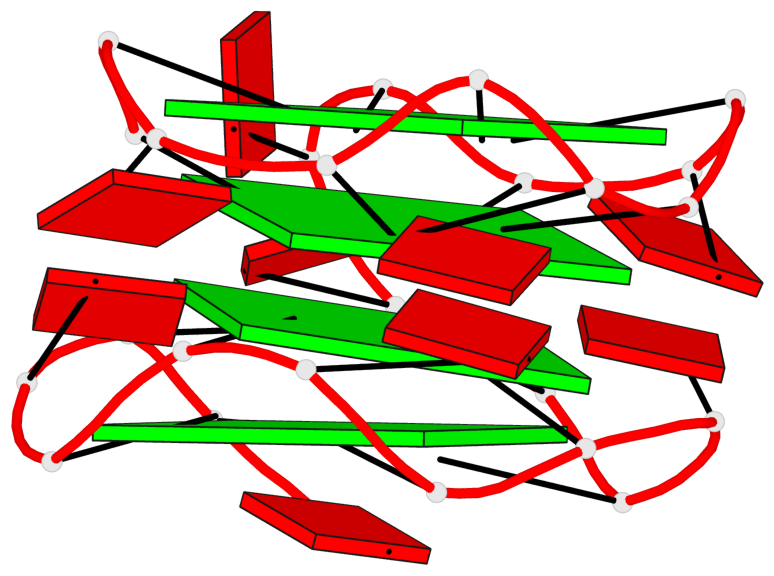
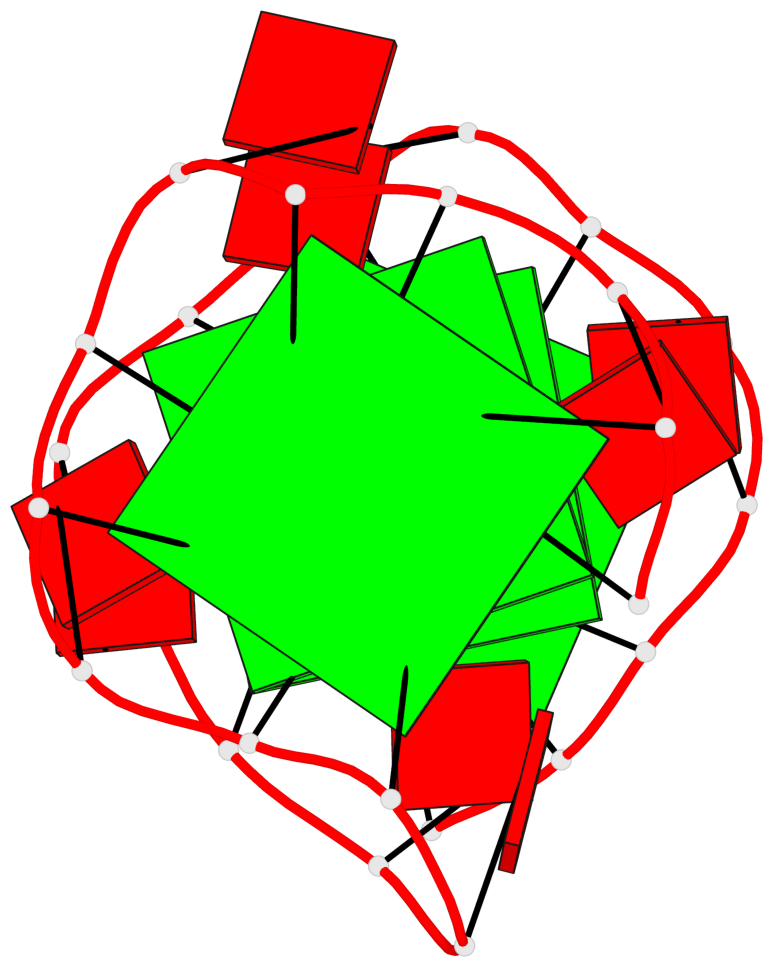
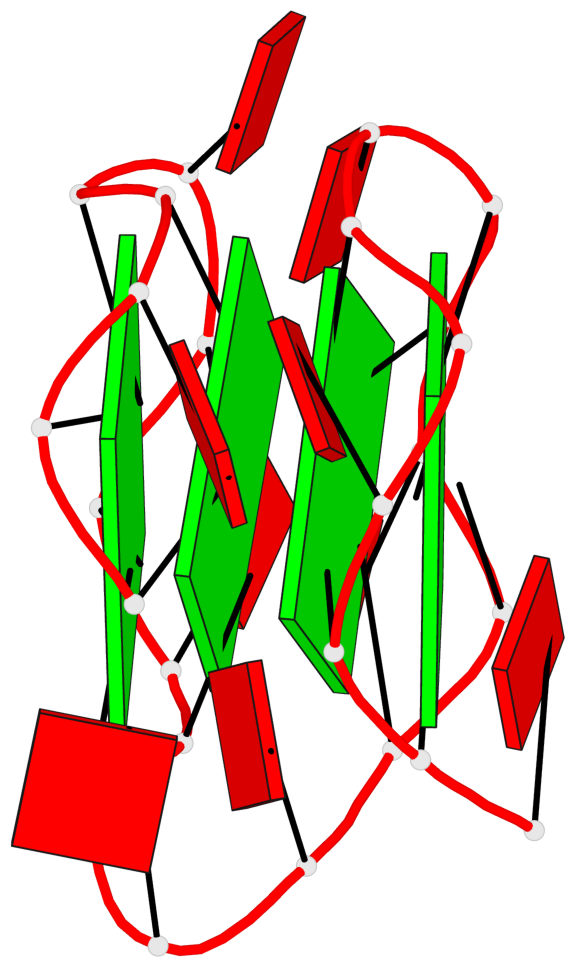
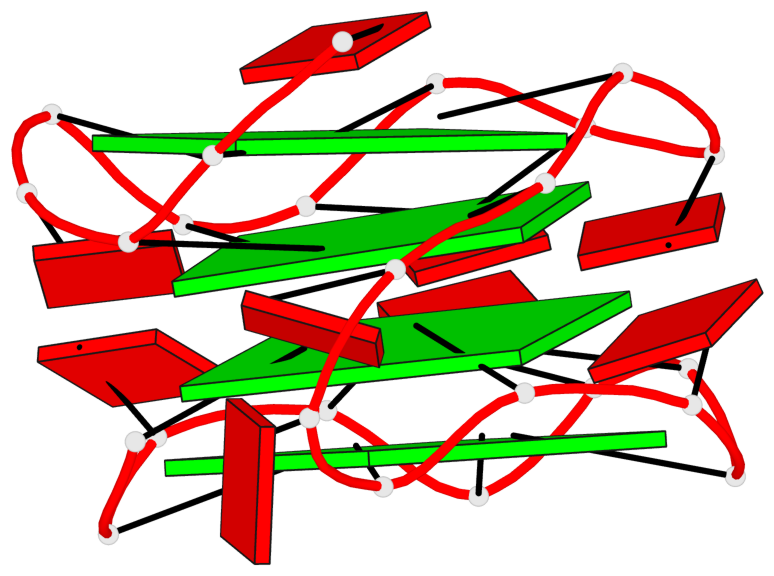
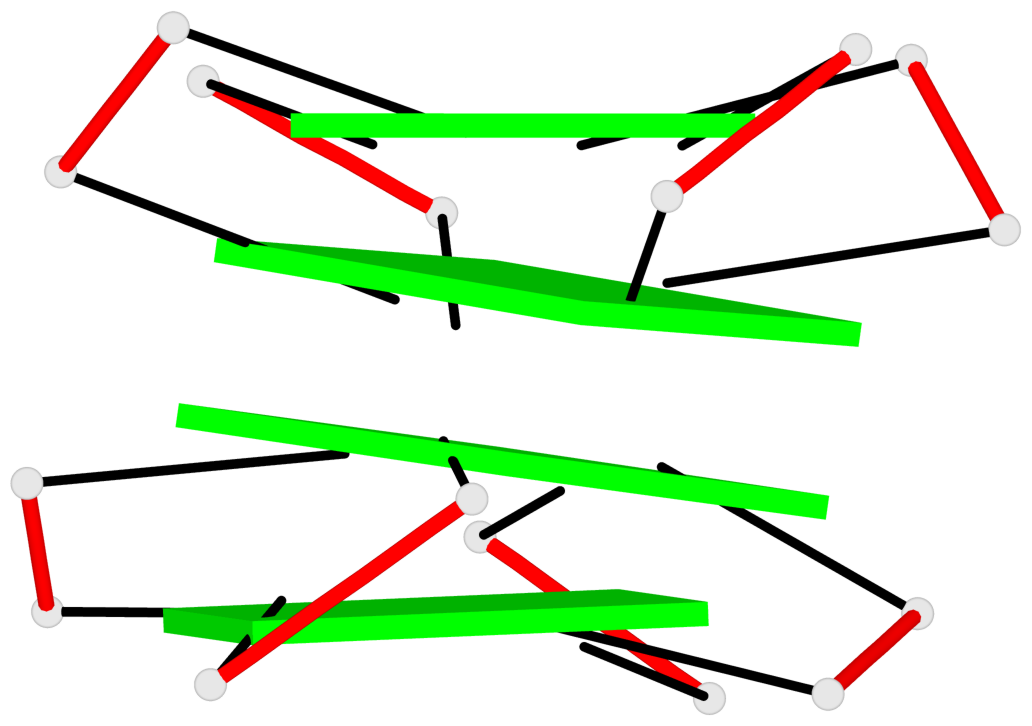
Base-block schematics in six views
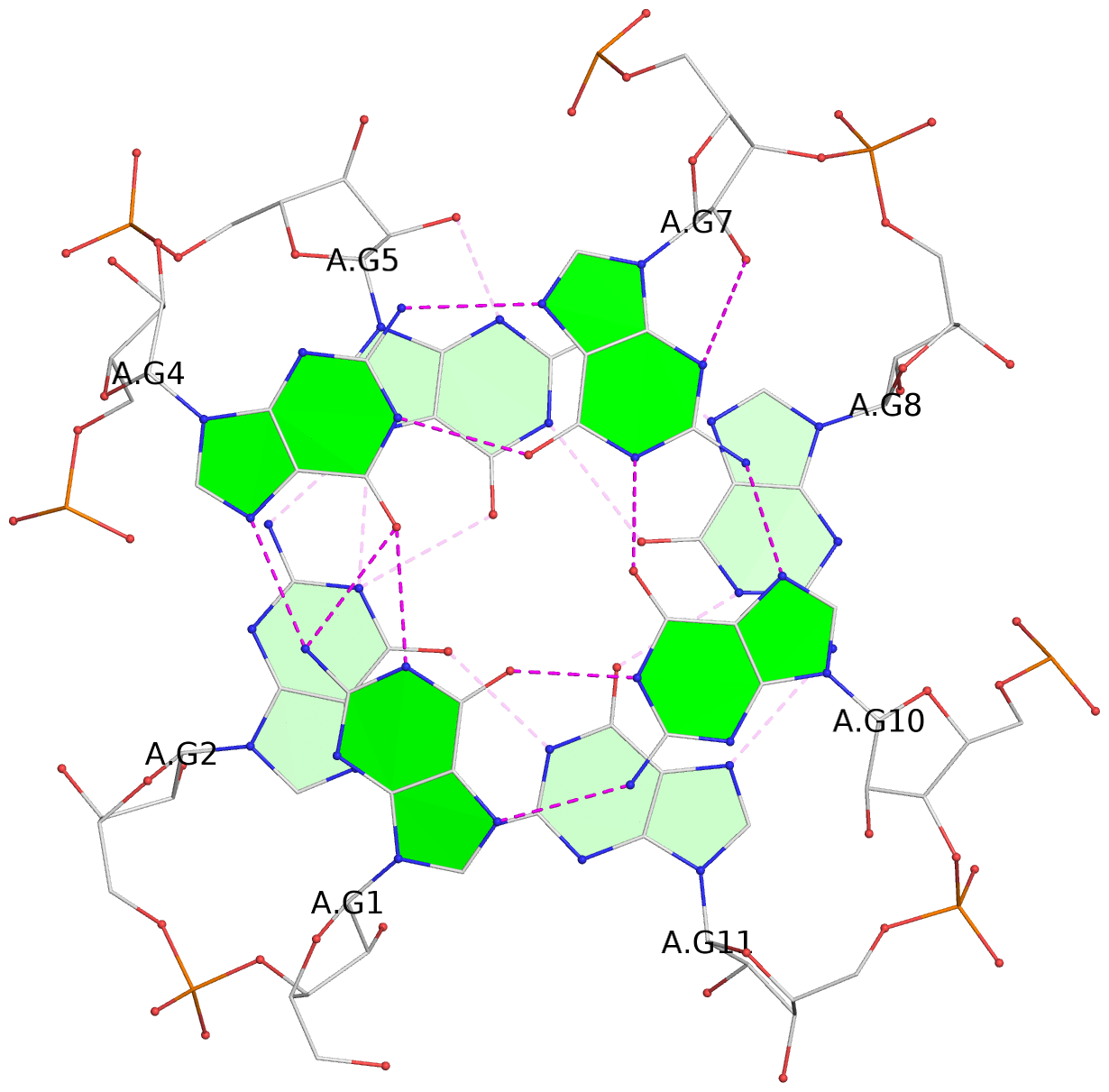
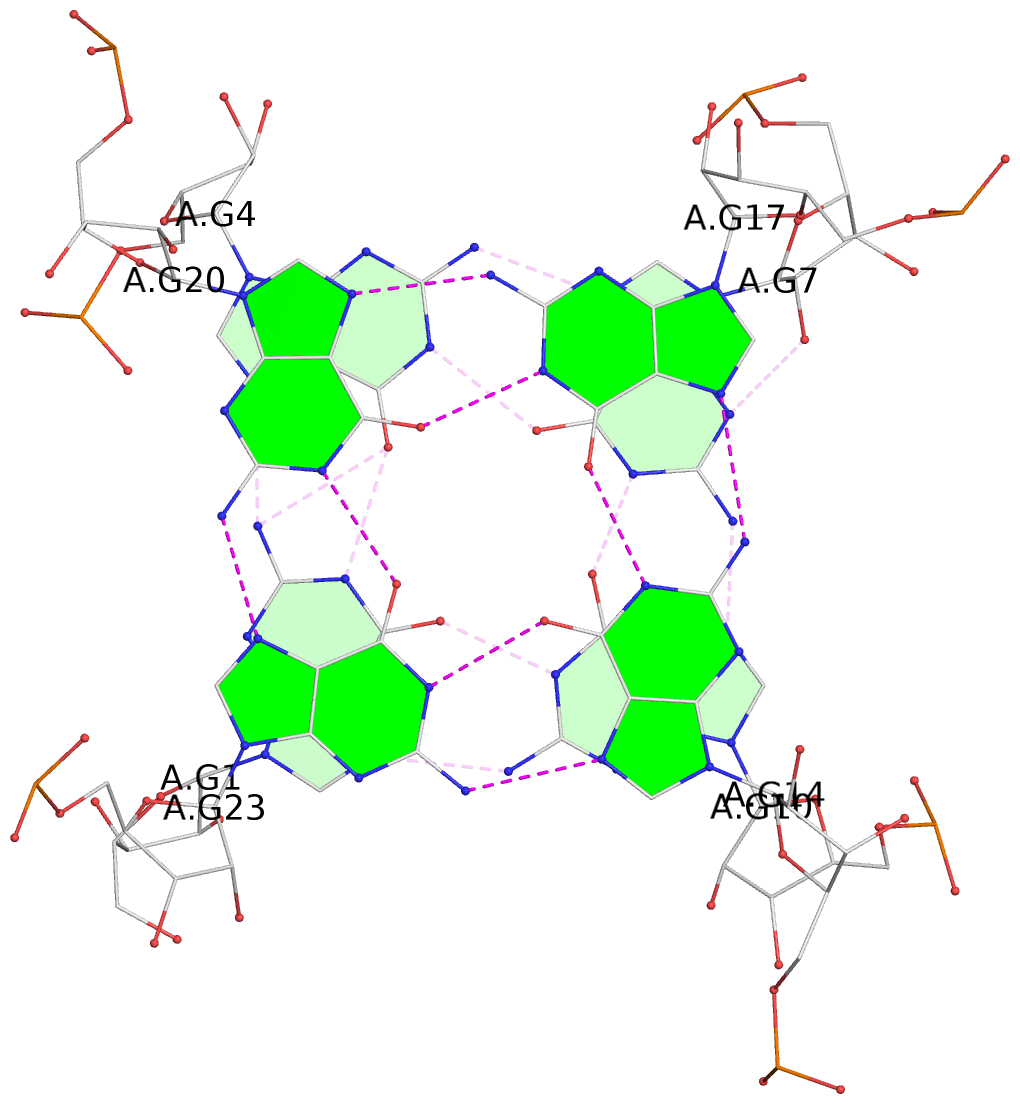
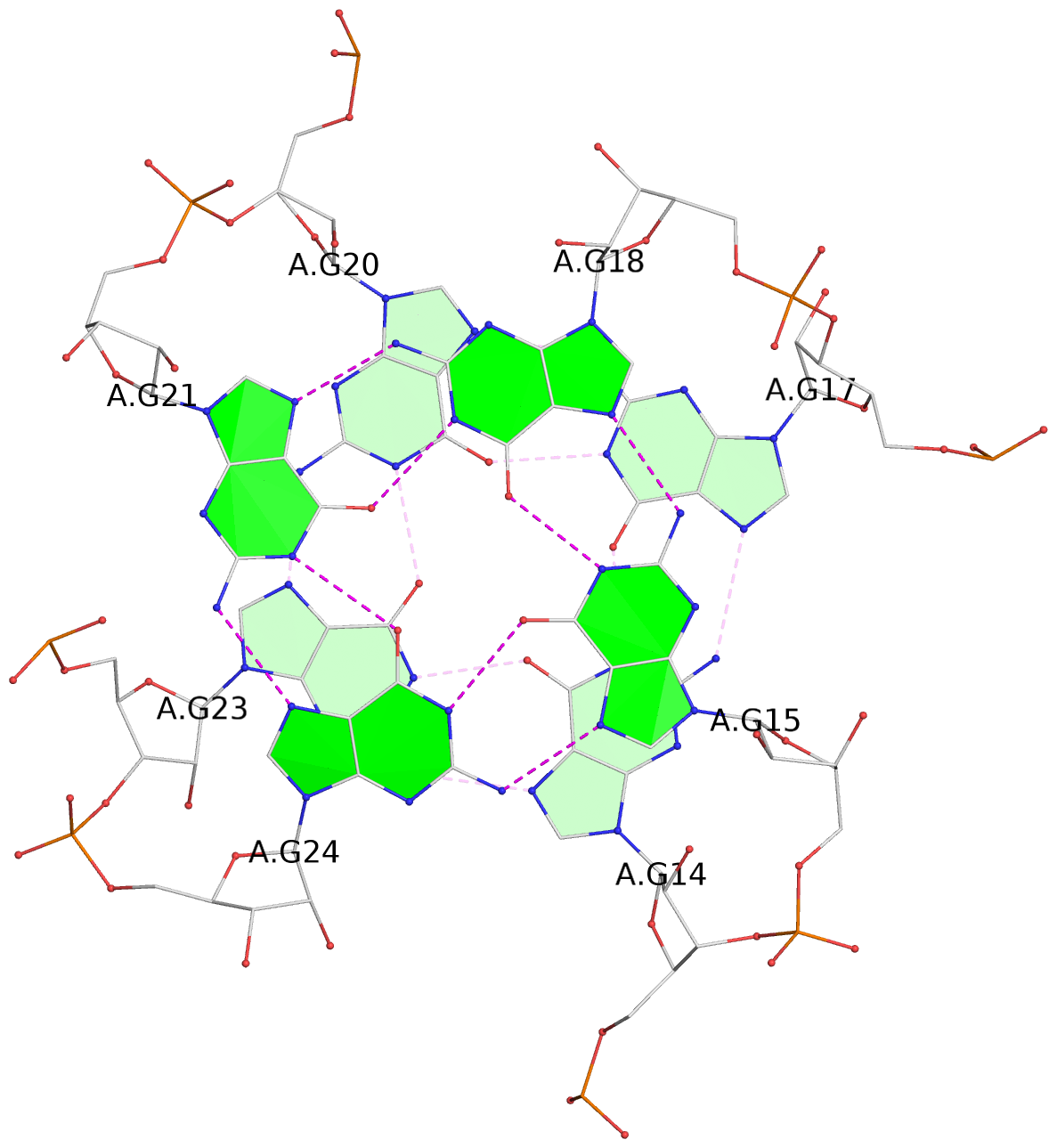
List of 4 G-tetrads
1 glyco-bond=---- sugar=-3-. groove=---- planarity=0.364 type=bowl nts=4 GGGG A.G1,A.G4,A.G7,A.G10 2 glyco-bond=---- sugar=---- groove=---- planarity=0.385 type=bowl nts=4 GGGG A.G2,A.G5,A.G8,A.G11 3 glyco-bond=---- sugar=-3-. groove=---- planarity=0.193 type=other nts=4 GGGG A.G14,A.G17,A.G20,A.G23 4 glyco-bond=---- sugar=---3 groove=---- planarity=0.390 type=other nts=4 GGGG A.G15,A.G18,A.G21,A.G24
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 4 G-tetrad layers, INTRA-molecular, with 2 stems
List of 2 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(-P-P-P), parallel(4+0)
Stem#2, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(-P-P-P), parallel(4+0)
List of 1 G4 coaxial stack
1 G4 helix#1 contains 2 G4 stems: [#1,#2] [5'/5']