Detailed DSSR results for the G-quadruplex: PDB entry 6kn4
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 6kn4
- Class
- DNA
- Method
- NMR
- Summary
- Human parallel stranded 7-mer g-quadruplex complexed with 2 adriamycin (dm2) molecules
- Reference
- Barthwal R, Raje S, Pandav K (2021): "Structural basis for stabilization of human telomeric G-quadruplex [d-(TTAGGGT)] 4 by anticancer drug adriamycin." J.Biomol.Struct.Dyn., 39, 795-815. doi: 10.1080/07391102.2020.1730969.
- Abstract
- Besides inhibiting DNA duplication, DNA dependent RNA synthesis and topoisomerase-II enzyme action, anticancer drug adriamycin is found to cause telomere dysfunction and shows multiple strategies of action on gene functioning. We present evidence of binding of adriamycin to parallel stranded intermolecular [d-(TTAGGGT)]4 G-quadruplex DNA comprising human telomeric DNA by proton and phosphorus-31 nuclear magnetic resonance spectroscopy. Diffusion ordered spectroscopy shows formation of complex between the two molecules. Changes in chemical shift and line broadening of DNA and adriamycin protons suggest participation of specific chemical groups/moieties in interaction. Presence of sequential nuclear Overhauser enhancements at all base quartet steps and absence of large downfield shifts in 31P resonances give clear proof of absence of intercalation of adriamycin chromophore between base quartets. Restrained molecular dynamics simulations using observed 15 short intermolecular inter proton distance contacts depict stacking of ring D of adriamycin with terminal G6 quartet by displacing T7 base and external groove binding close to T1-T2-A3 bases. The disappearance of imino protons monitored as a function of temperature and differential scanning calorimetry experiments yield thermal stabilization of 24 °C, which is likely to come in the way of telomerase association with telomeres. The findings pave the way for design of alternate anthracycline based drugs with specific modifications at ring D to enhance induced thermal stabilization and use alternate mechanism of binding to G-quadruplex DNA for interference in functional pathway of telomere maintenance by telomerase enzyme besides their well known action on duplex DNA. Communicated by Ramaswamy H. Sarma.
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, parallel(4+0), UUUU
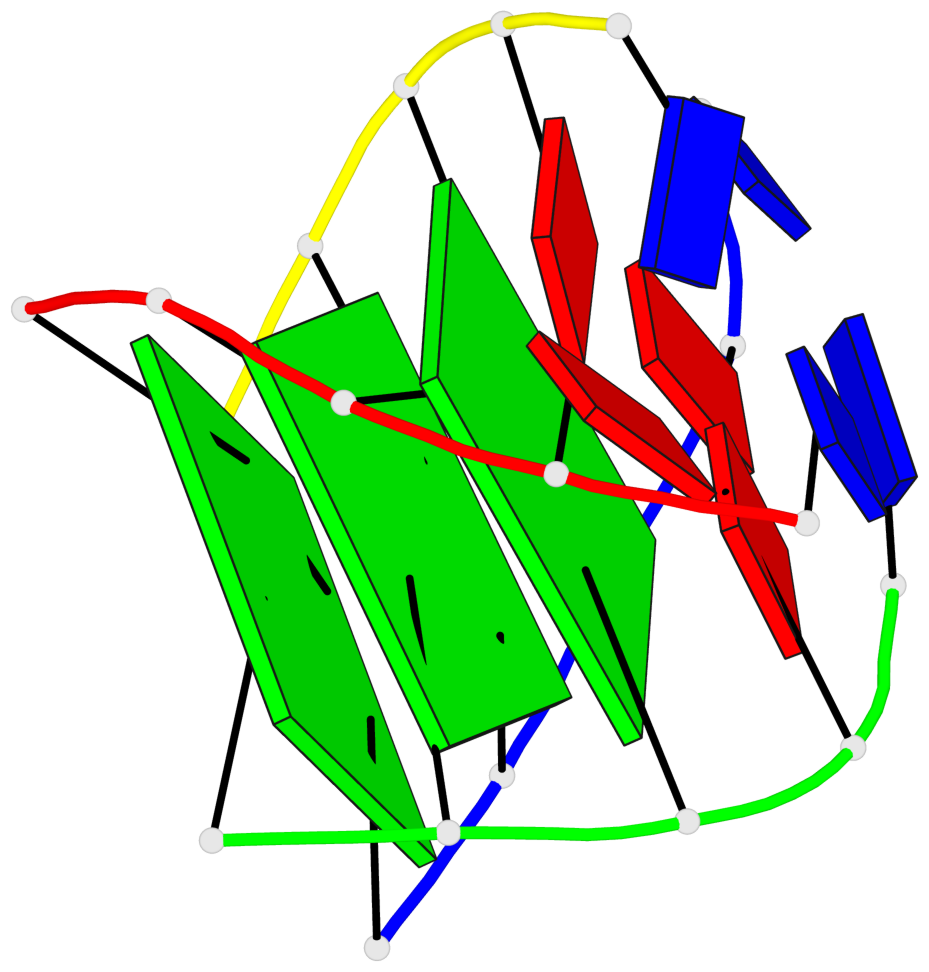
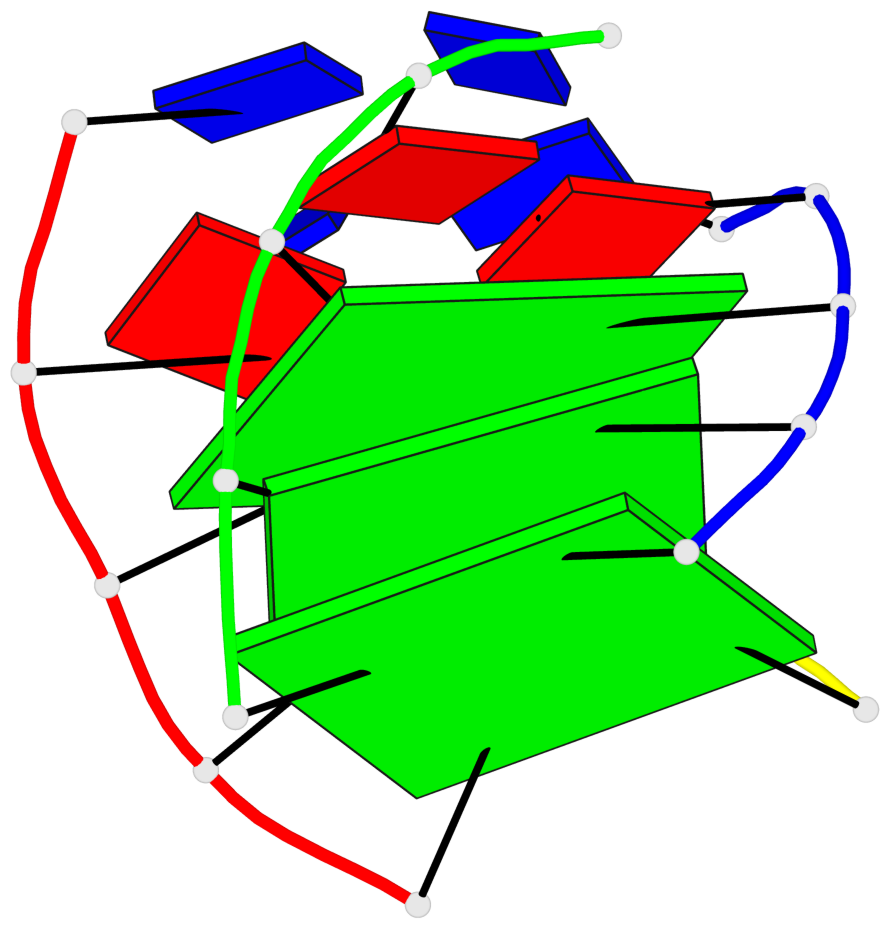
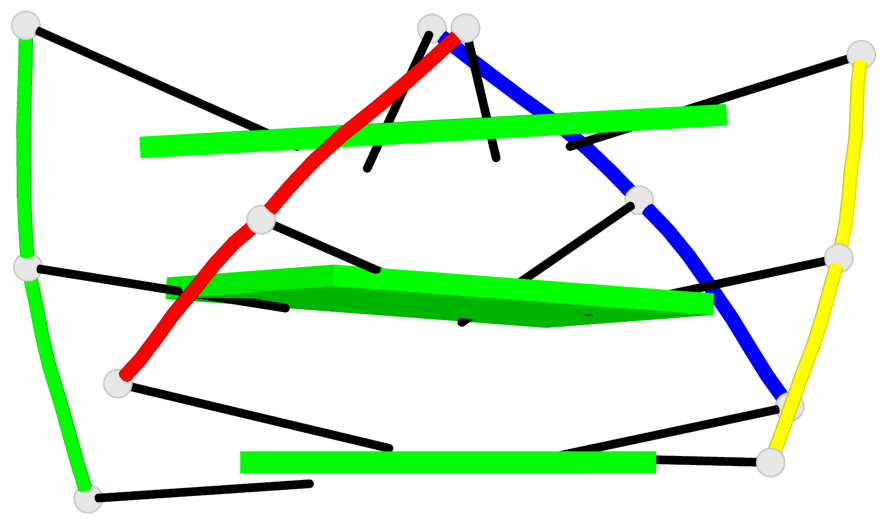
Base-block schematics in six views
List of 3 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.425 type=other nts=4 GGGG A.DG4,D.DG25,C.DG18,B.DG11 2 glyco-bond=---- sugar=.... groove=---- planarity=0.502 type=bowl nts=4 GGGG A.DG5,D.DG26,C.DG19,B.DG12 3 glyco-bond=---- sugar=.-.. groove=---- planarity=0.629 type=bowl nts=4 GGGG A.DG6,D.DG27,C.DG20,B.DG13
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, inter-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.