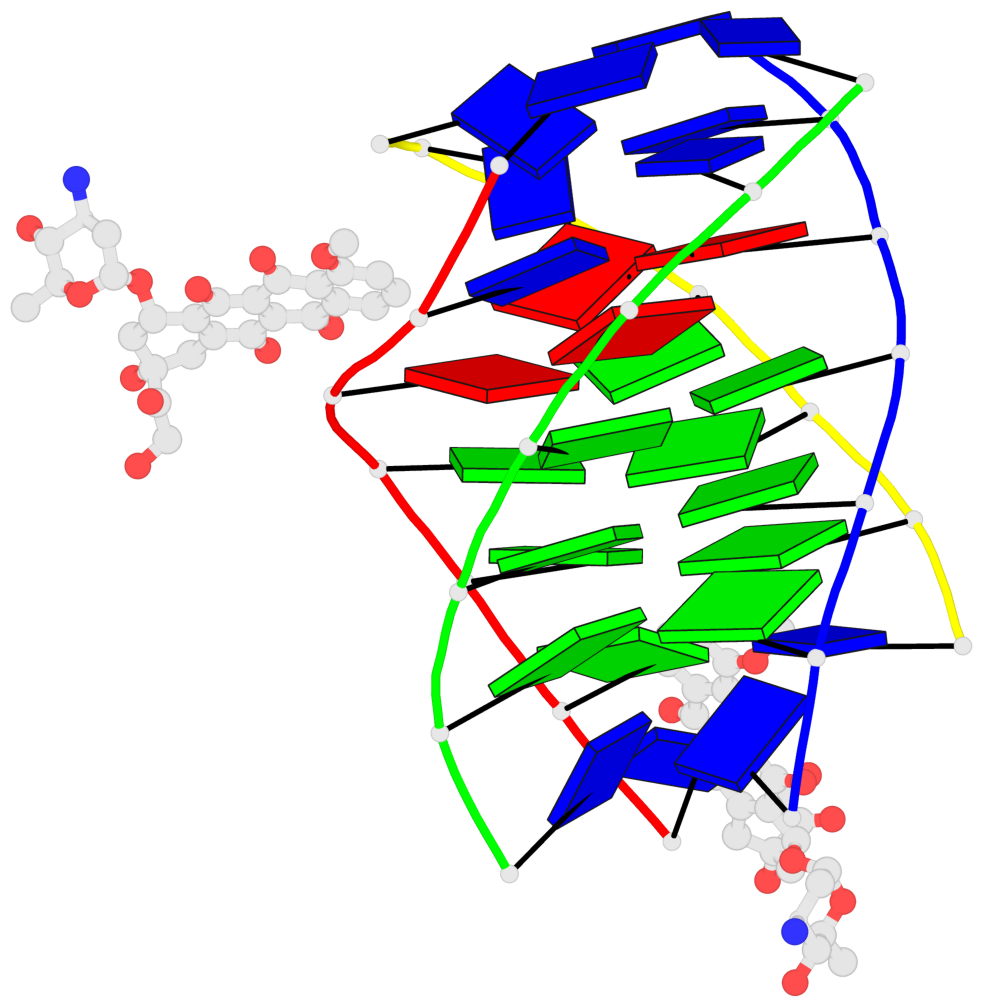
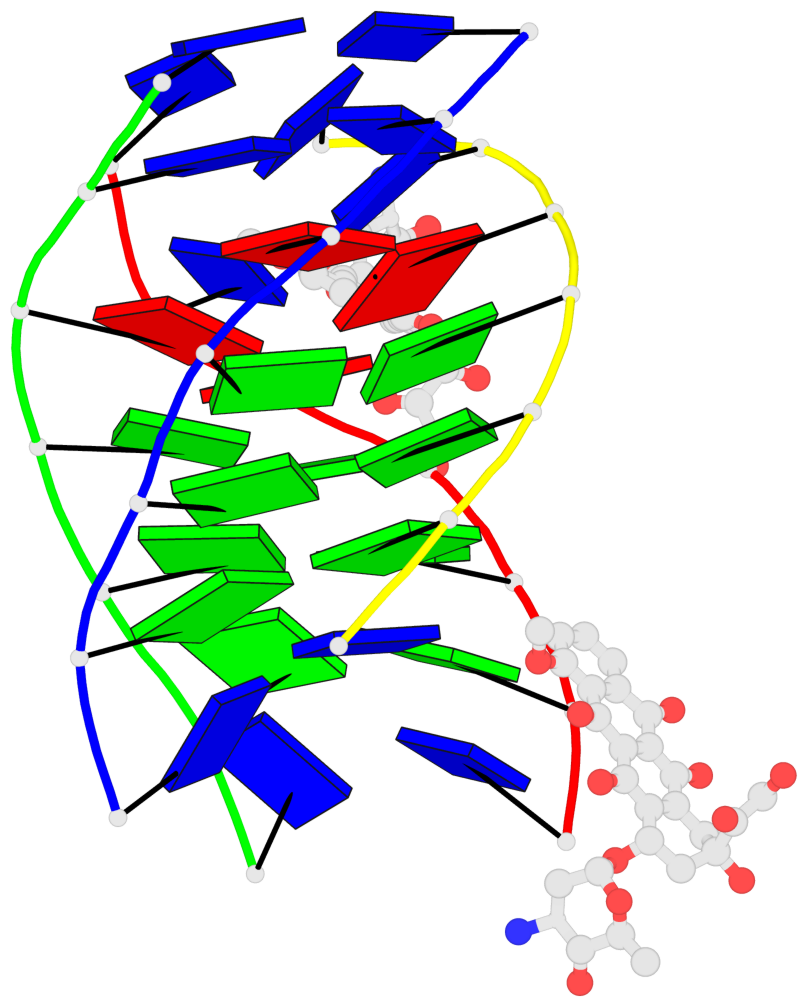
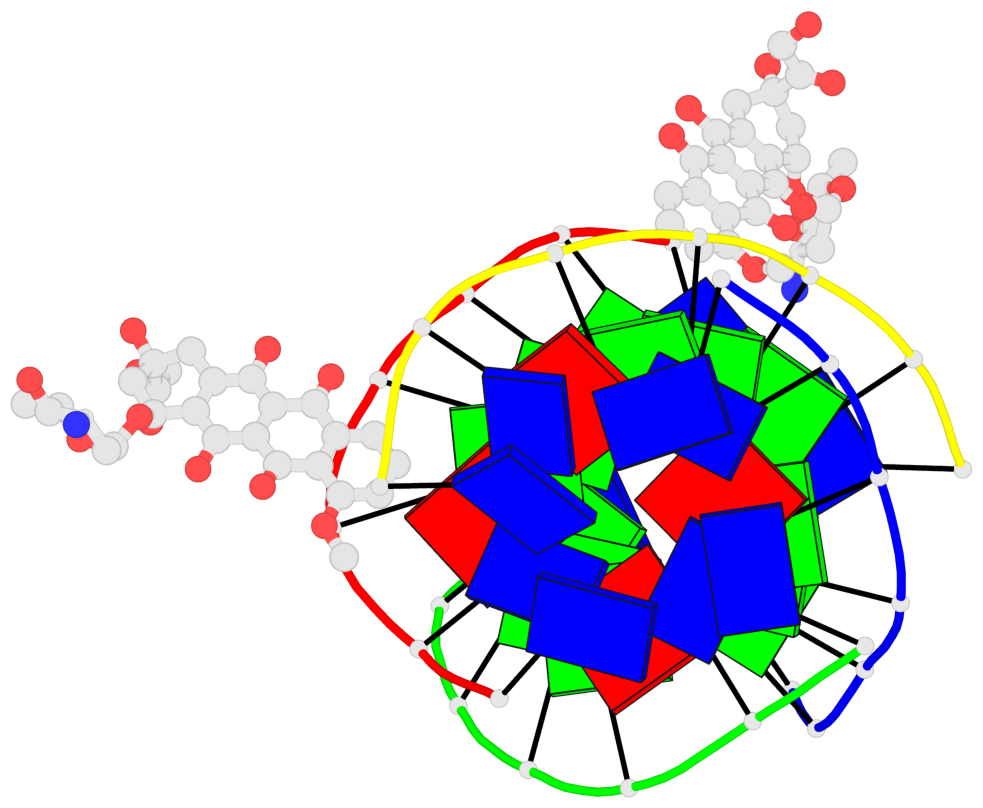
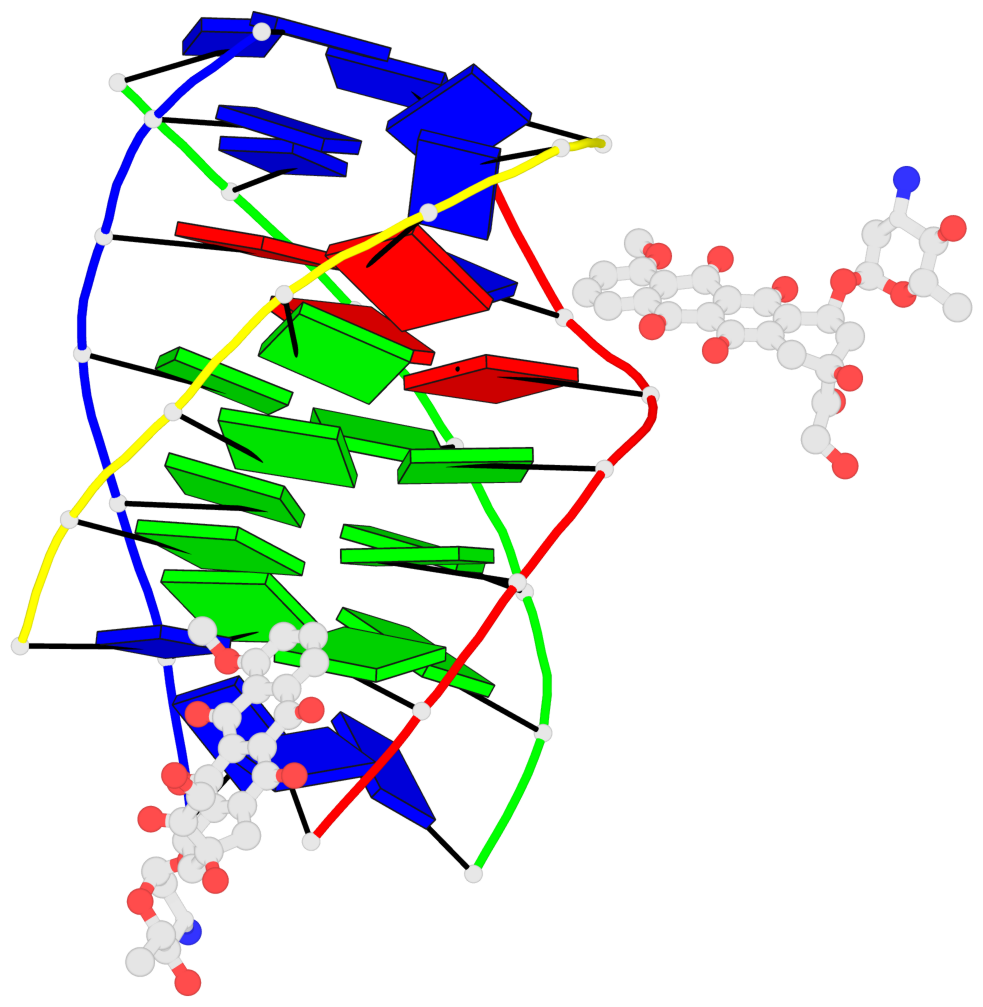
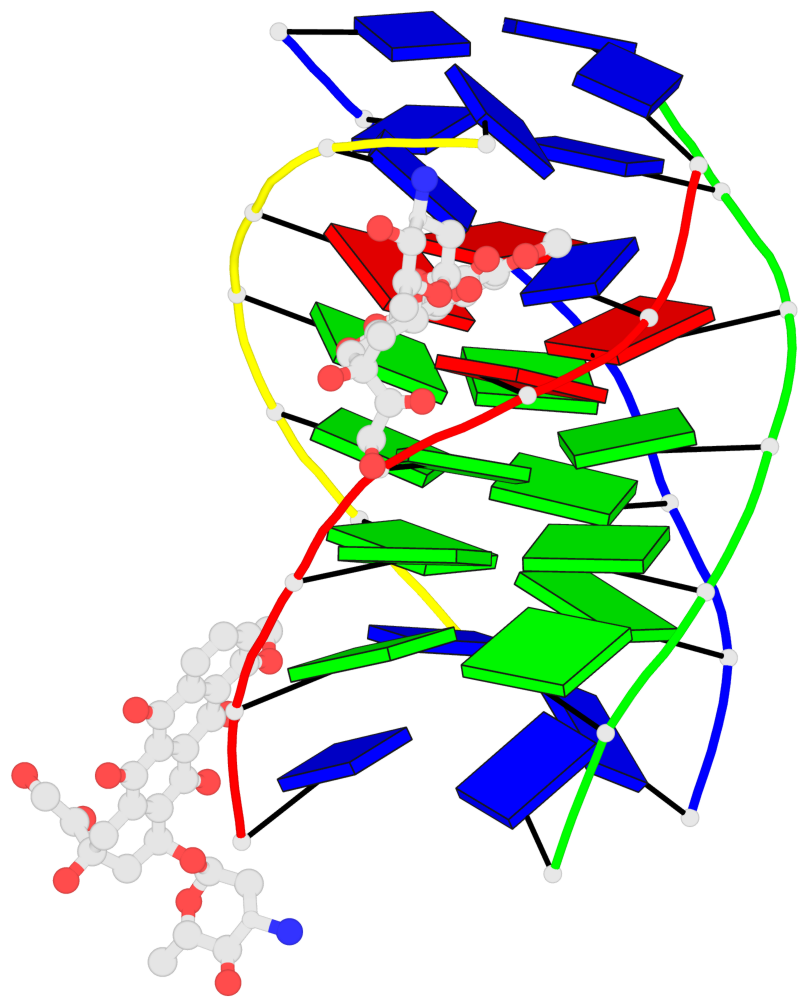
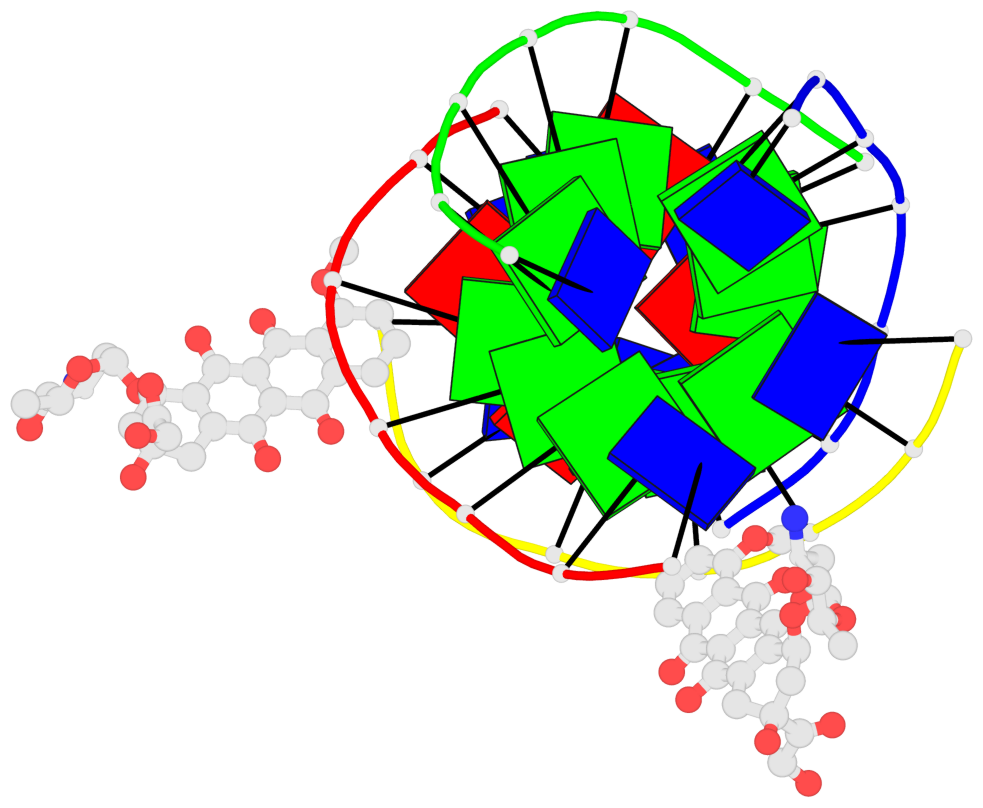
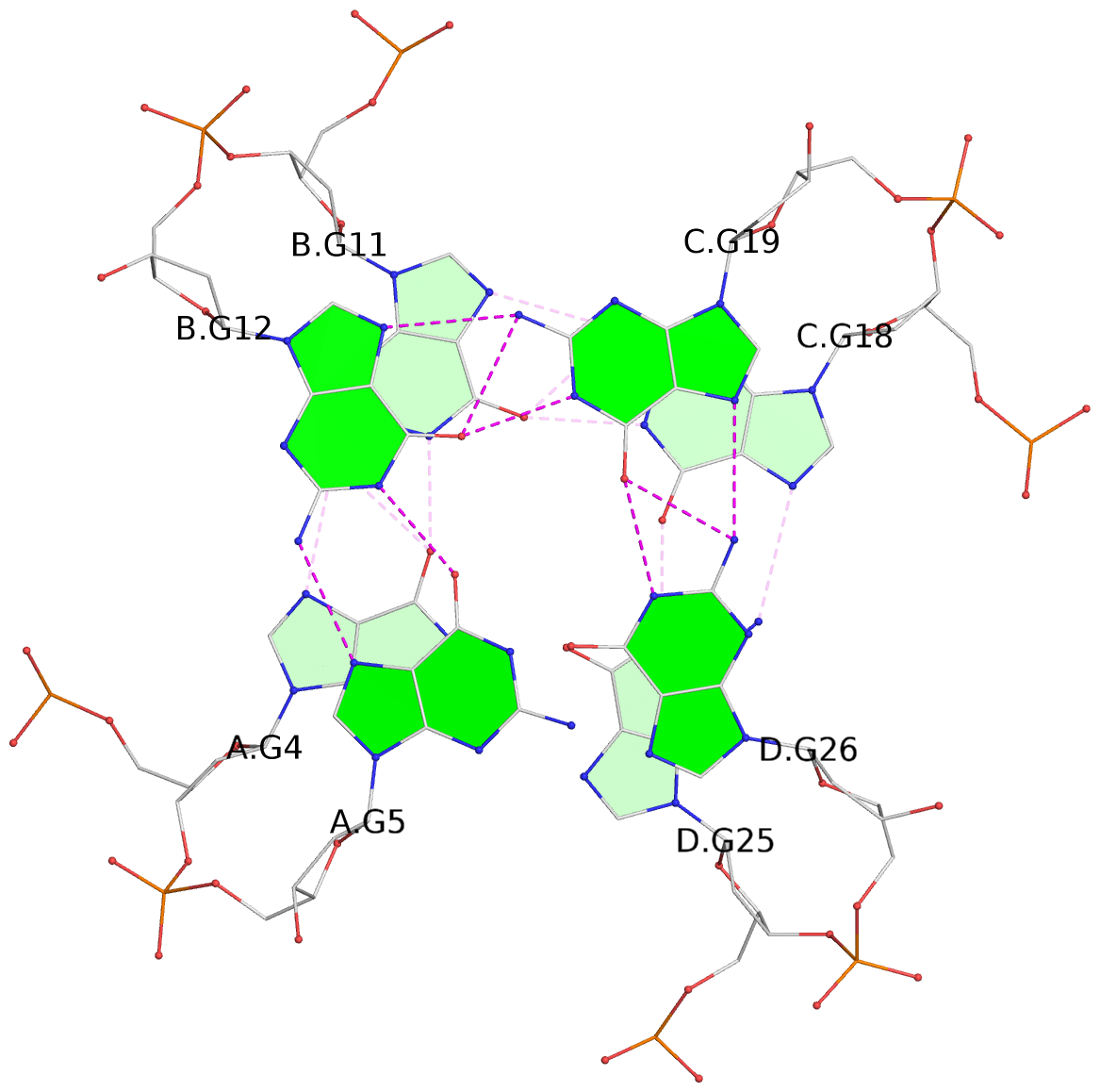
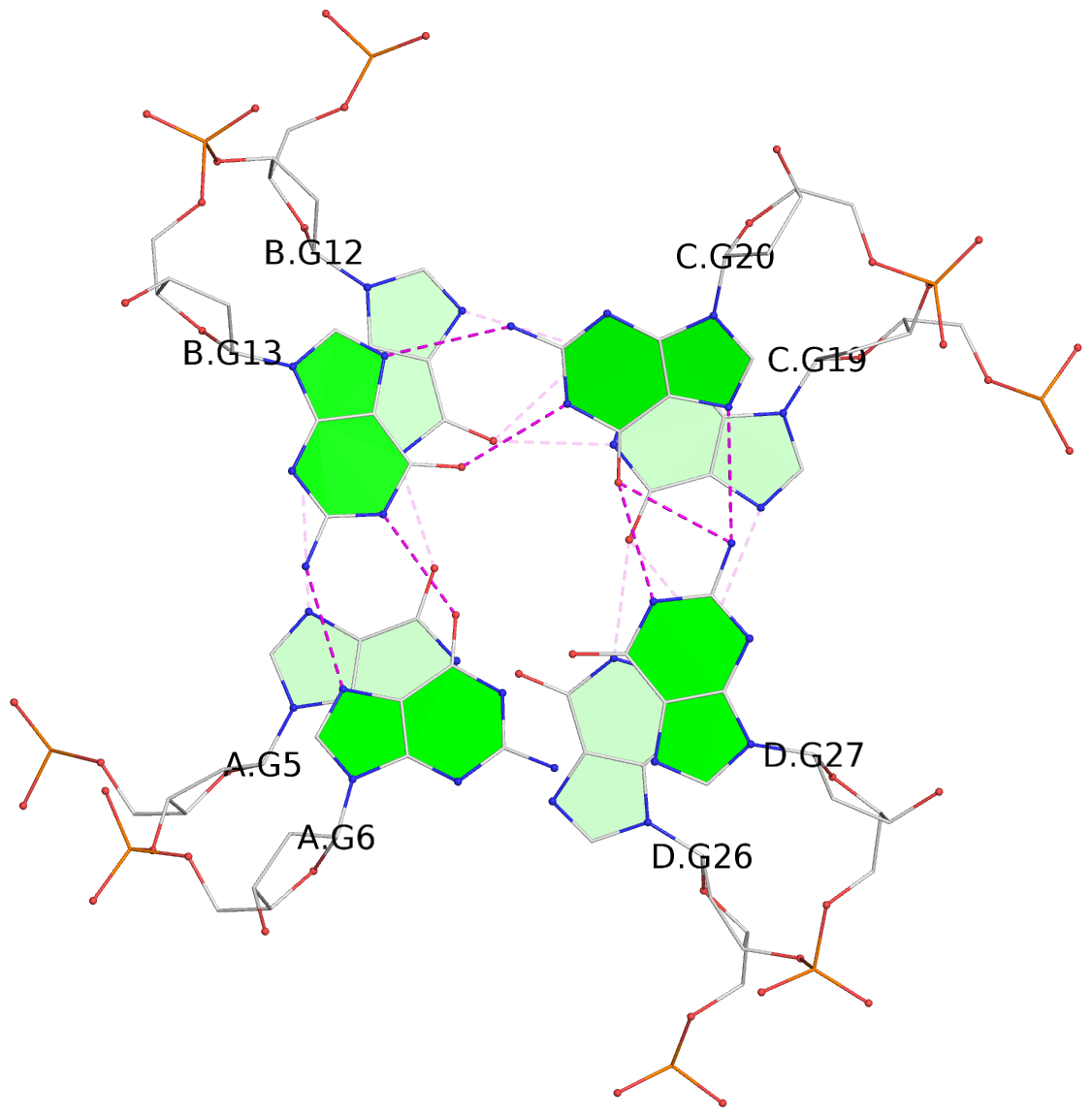
Detailed DSSR results for the G-quadruplex: PDB entry 6kxz
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Poster "DSSR-Enabled Automatic
Identification and Annotation of G-quadruplexes in the PDB"
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper,
or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 6kxz
- Class
- DNA
- Method
- NMR
- Summary
- Human parallel stranded 7-mer g-quadruplex complexed with 2 epirubicin (epi) molecules
- Reference
- Barthwal R, Raje S, Pandav K (2020): "Structural basis for stabilization of human telomeric G-quadruplex [d-(TTAGGGT)] 4 by anticancer drug epirubicin." Bioorg.Med.Chem., 28, 115761. doi: 10.1016/j.bmc.2020.115761.
- Abstract
- Anthracycline anticancer drugs show multiple strategies of action on gene functioning by regulation of telomerase enzyme by apoptotic factors, e.g. ceramide level, p53 activity, bcl-2 protein levels, besides inhibiting DNA/RNA synthesis and topoisomerase-II action. We report binding of epirubicin with G-quadruplex (G4) DNA, [d-(TTAGGGT)]4, comprising human telomeric DNA sequence TTAGGG, using 1H and 31P NMR spectroscopy. Diffusion ordered spectroscopy, sequence selective changes in chemical shift (~0.33 ppm) and line broadening in DNA signals suggest formation of a well-defined complex. Presence of sequential nuclear Overhauser enhancements at all base quartet steps and absence of large downfield shifts in 31P resonances preclude intercalative mode of interaction. Restrained molecular dynamics simulations using AMBER force field incorporating intermolecular drug to DNA interproton distances, involving ring D protons of epirubicin depict external binding close to T1-T2-A3 and G6pT7 sites. Binding induced thermal stabilization of G4 DNA (~36 °C), obtained from imino protons and differential scanning calorimetry, is likely to come in the way of telomerase association with telomeres. The findings pave the way for drug-designing with modifications at ring D and daunosamine sugar.
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, parallel(4+0), UUUU
Base-block schematics in six views
List of 3 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.803 type=other nts=4 GGGG A.DG4,B.DG11,C.DG18,D.DG25 2 glyco-bond=---- sugar=.-.. groove=---- planarity=0.796 type=other nts=4 GGGG A.DG5,B.DG12,C.DG19,D.DG26 3 glyco-bond=---- sugar=---- groove=---- planarity=0.859 type=other nts=4 GGGG A.DG6,B.DG13,C.DG20,D.DG27
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, inter-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.