Detailed DSSR results for the G-quadruplex: PDB entry 6l8m
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 6l8m
- Class
- DNA
- Method
- NMR
- Summary
- Wnt DNA promoter mutant g-quadruplex
- Reference
- Wang ZF, Li MH, Chu IT, Winnerdy FR, Phan AT, Chang TC (2020): "Cytosine epigenetic modification modulates the formation of an unprecedented G4 structure in the WNT1 promoter." Nucleic Acids Res., 48, 1120-1130. doi: 10.1093/nar/gkz1207.
- Abstract
- Time-resolved imino proton nuclear magnetic resonance spectra of the WT22m sequence d(GGGCCACCGGGCAGTGGGCGGG), derived from the WNT1 promoter region, revealed an intermediate G-quadruplex G4(I) structure during K+-induced conformational transition from an initial hairpin structure to the final G4(II) structure. Moreover, a single-base C-to-T mutation at either position C4 or C7 of WT22m could lock the intermediate G4(I) structure without further conformational change to the final G4(II) structure. Surprisingly, we found that the intermediate G4(I) structure is an atypical G4 structure, which differs from a typical hybrid G4 structure of the final G4(II) structure. Further studies of modified cytosine analogues associated with epigenetic regulation indicated that slight modification on a cytosine could modulate G4 structure. A simplified four-state transition model was introduced to describe such conformational transition and disclose the possible mechanism for G4 structural selection caused by cytosine modification.
- G4 notes
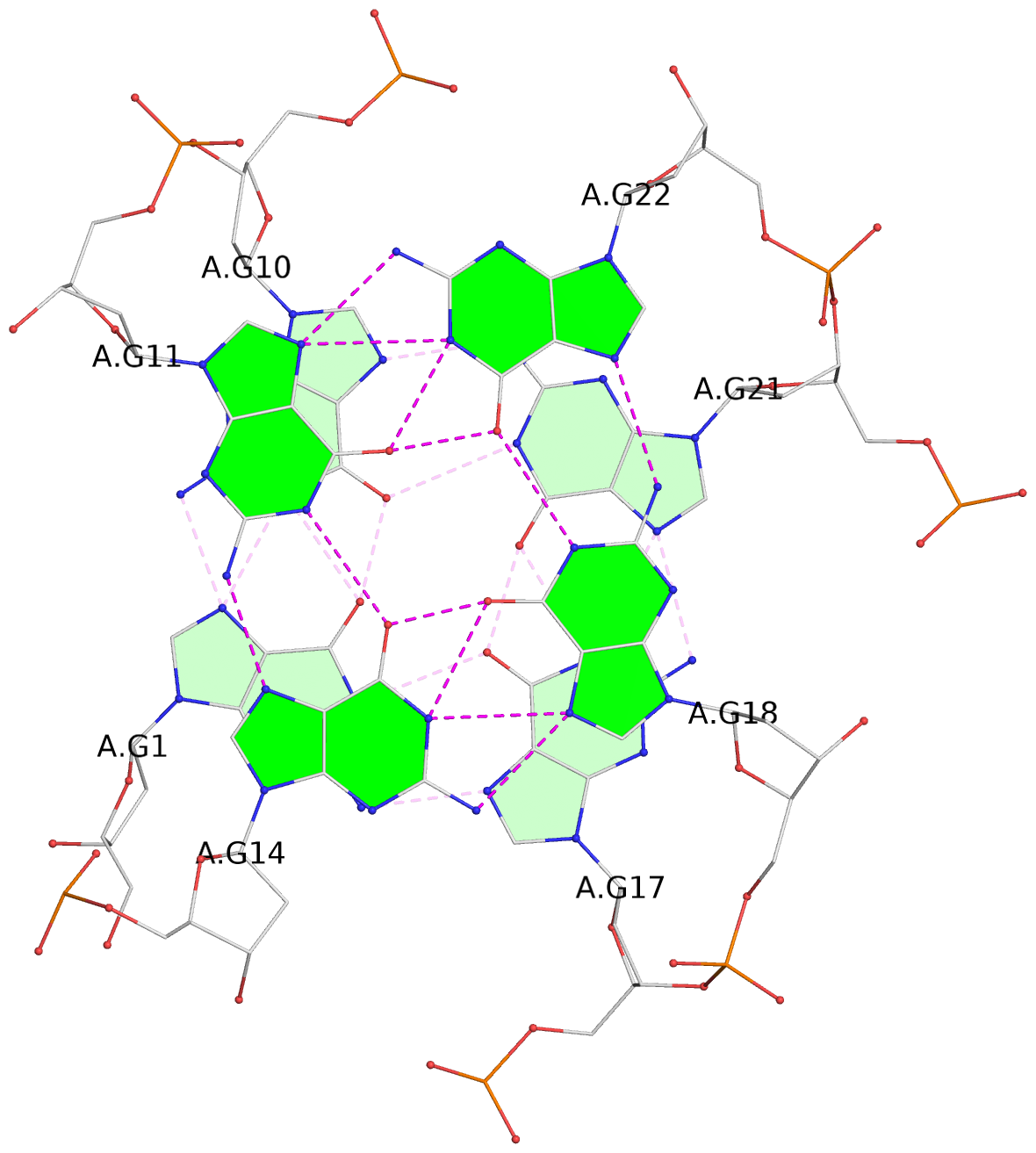
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 2(-LwX+P), UD3(1+3), UDDD
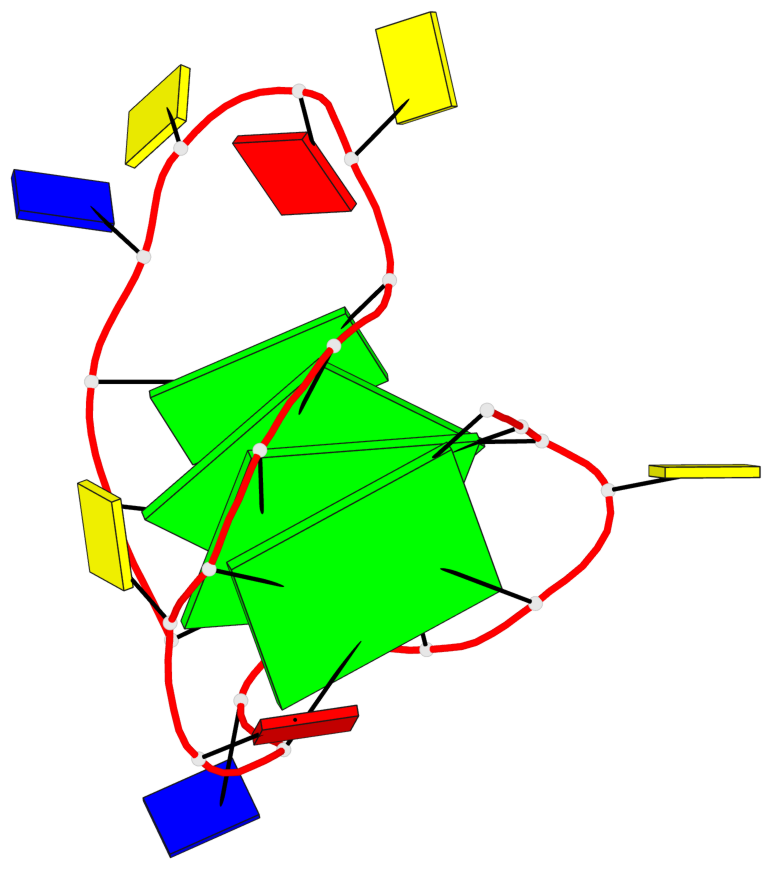
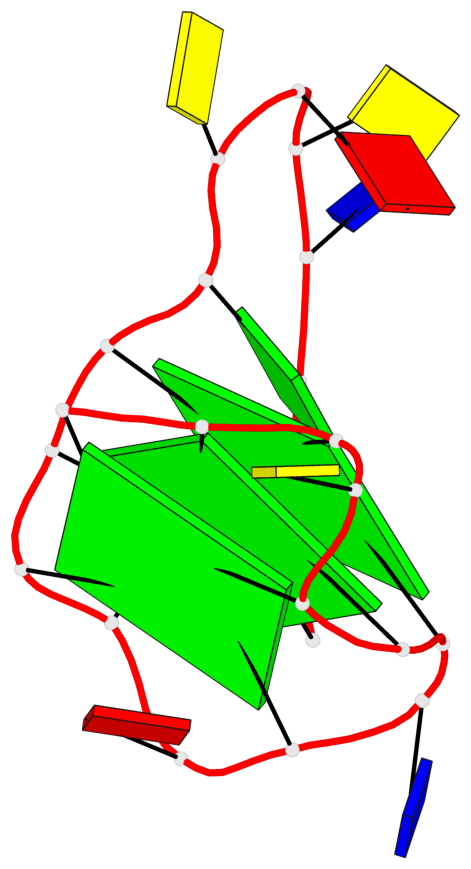
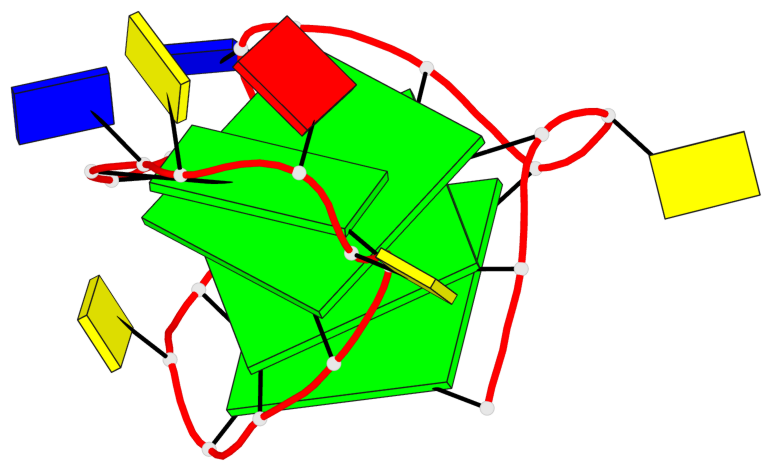
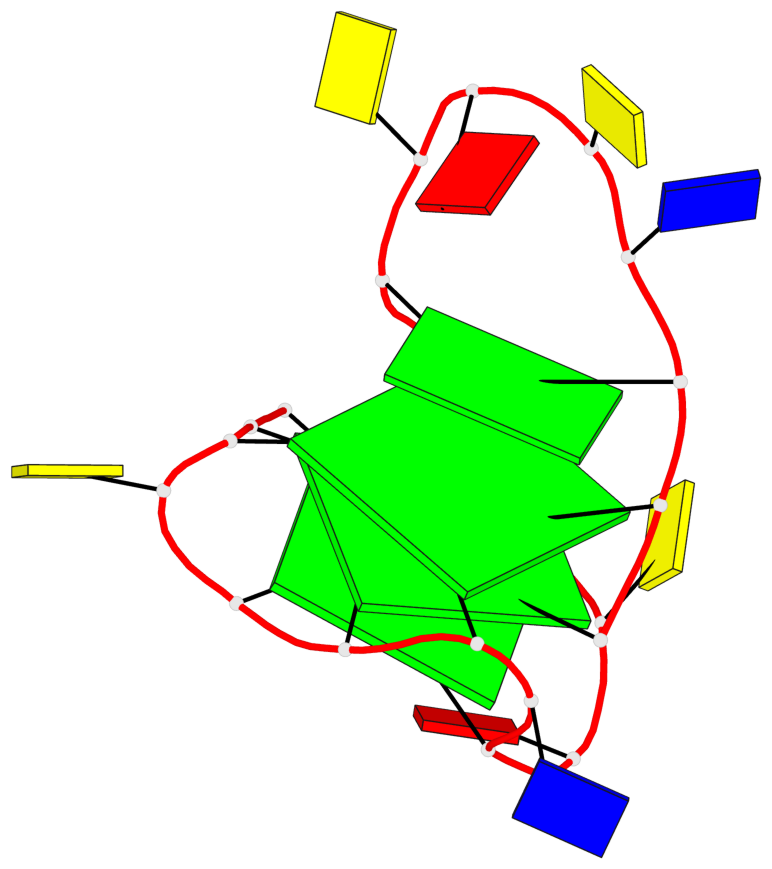
Base-block schematics in six views
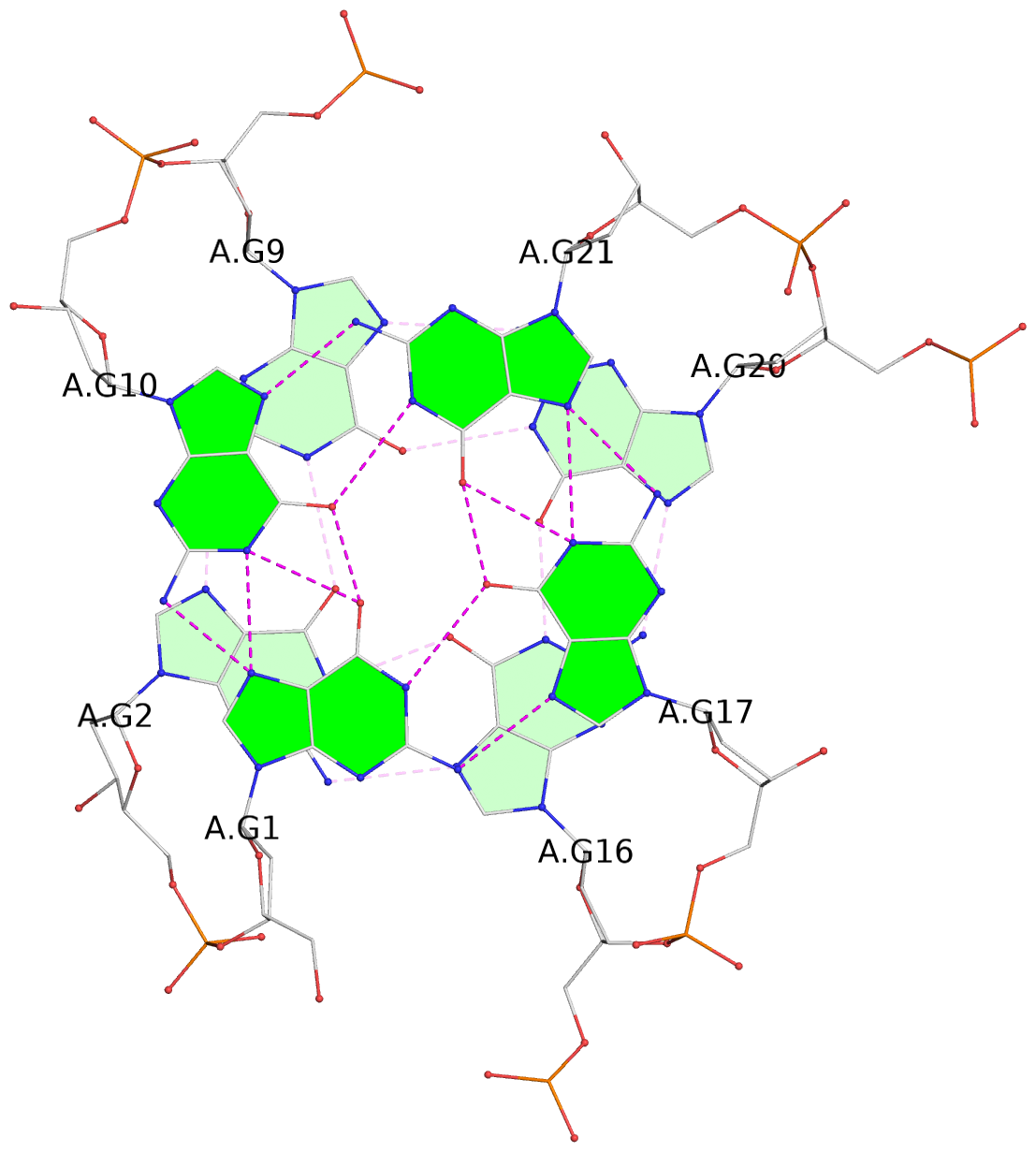
List of 3 G-tetrads
1 glyco-bond=s--- sugar=-.-- groove=w--n planarity=0.061 type=planar nts=4 GGGG A.DG1,A.DG10,A.DG21,A.DG17 2 glyco-bond=s--- sugar=3--- groove=w--n planarity=0.074 type=planar nts=4 GGGG A.DG2,A.DG9,A.DG20,A.DG16 3 glyco-bond=---- sugar=-.-- groove=---- planarity=0.076 type=planar nts=4 GGGG A.DG11,A.DG14,A.DG18,A.DG22
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 2 G-tetrad layers, 3 loops, INTRA-molecular, UDDD, hybrid-(mixed), 2(-LwX+P), UD3(1+3)
List of 2 non-stem G4-loops (including the two closing Gs)
1 type=lateral helix=#1 nts=4 GCAG A.DG11,A.DC12,A.DA13,A.DG14 2 type=V-shaped helix=#1 nts=5 GTGGG A.DG14,A.DT15,A.DG16,A.DG17,A.DG18