Detailed DSSR results for the G-quadruplex: PDB entry 6ldm
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 6ldm
- Class
- DNA binding protein-DNA
- Method
- X-ray (2.4 Å)
- Summary
- Structural basis of g-quadruplex DNA recognition by the yeast telomeric protein rap1
- Reference
- Traczyk A, Liew CW, Gill DJ, Rhodes D (2020): "Structural basis of G-quadruplex DNA recognition by the yeast telomeric protein Rap1." Nucleic Acids Res., 48, 4562-4571. doi: 10.1093/nar/gkaa171.
- Abstract
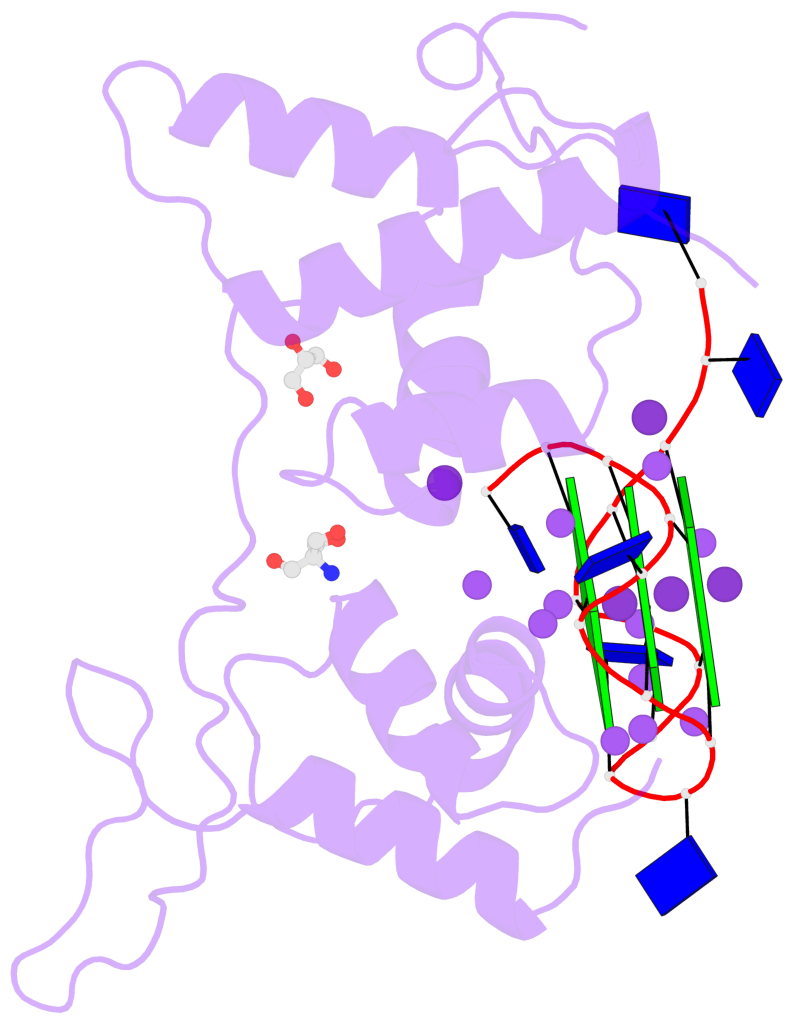
- G-quadruplexes are four-stranded nucleic acid structures involved in multiple cellular pathways including DNA replication and telomere maintenance. Such structures are formed by G-rich DNA sequences typified by telomeric DNA repeats. Whilst there is evidence for proteins that bind and regulate G-quadruplex formation, the molecular basis for this remains poorly understood. The budding yeast telomeric protein Rap1, originally identified as a transcriptional regulator functioning by recognizing double-stranded DNA binding sites, was one of the first proteins to be discovered to also bind and promote G-quadruplex formation in vitro. Here, we present the 2.4 Å resolution crystal structure of the Rap1 DNA-binding domain in complex with a G-quadruplex. Our structure not only provides a detailed insight into the structural basis for G-quadruplex recognition by a protein, but also gives a mechanistic understanding of how the same DNA-binding domain adapts to specifically recognize different DNA structures. The key observation is the DNA-recognition helix functions in a bimodal manner: In double-stranded DNA recognition one helix face makes electrostatic interactions with the major groove of DNA, whereas in G-quadruplex recognition a different helix face is used to make primarily hydrophobic interactions with the planar face of a G-tetrad.
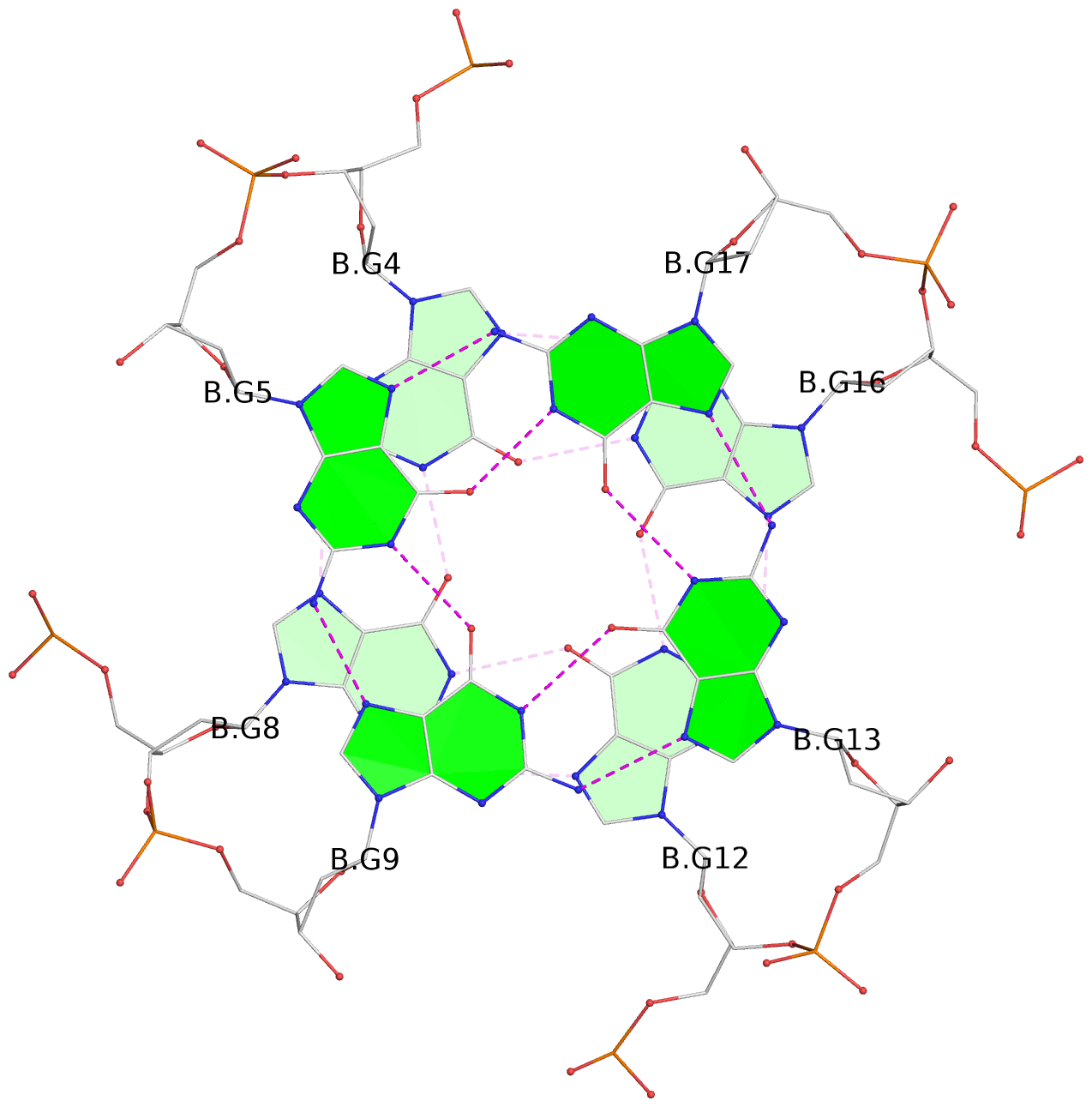
- G4 notes
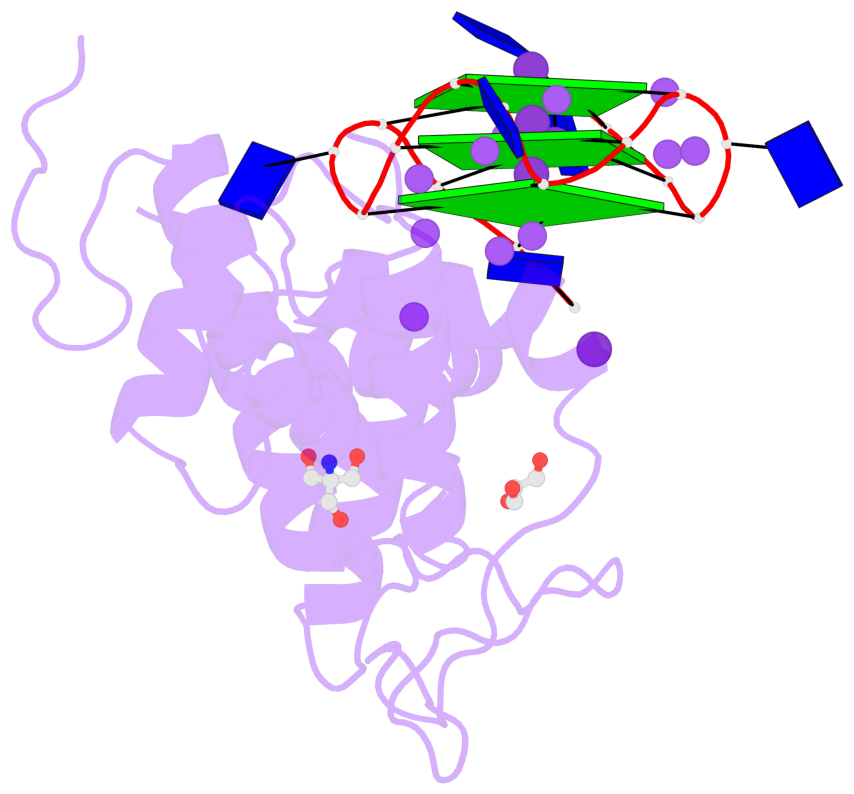
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-P-P-P), parallel(4+0), UUUU
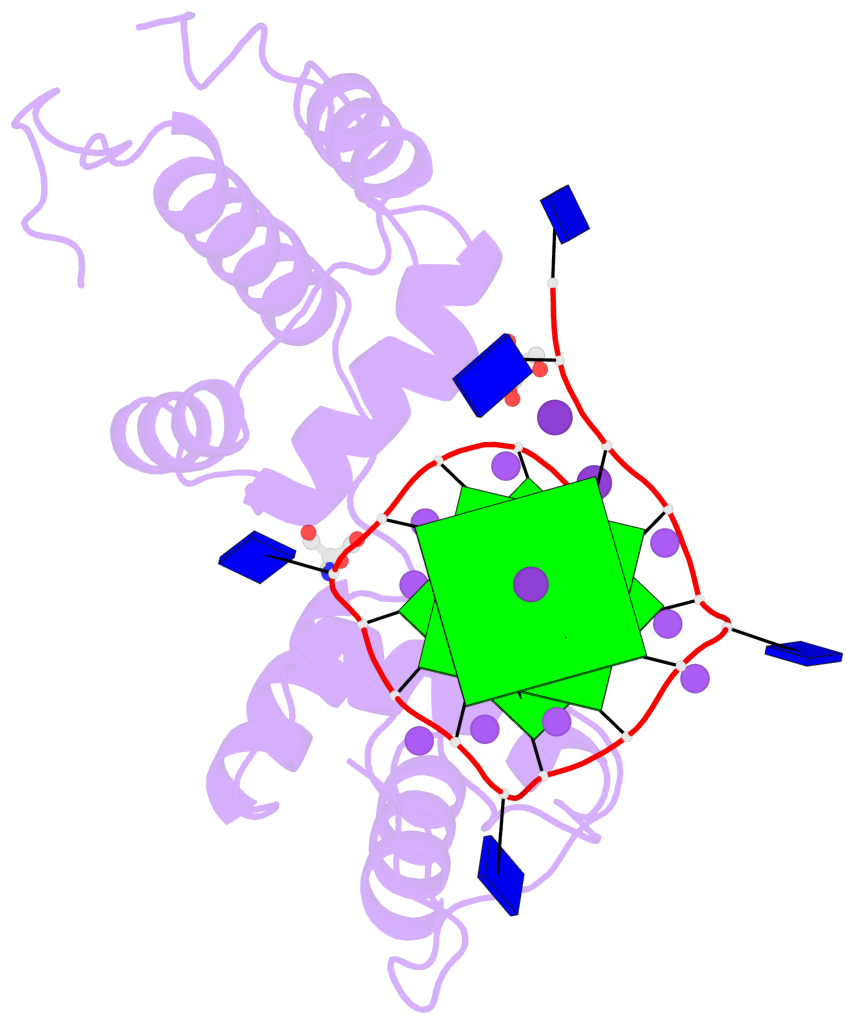
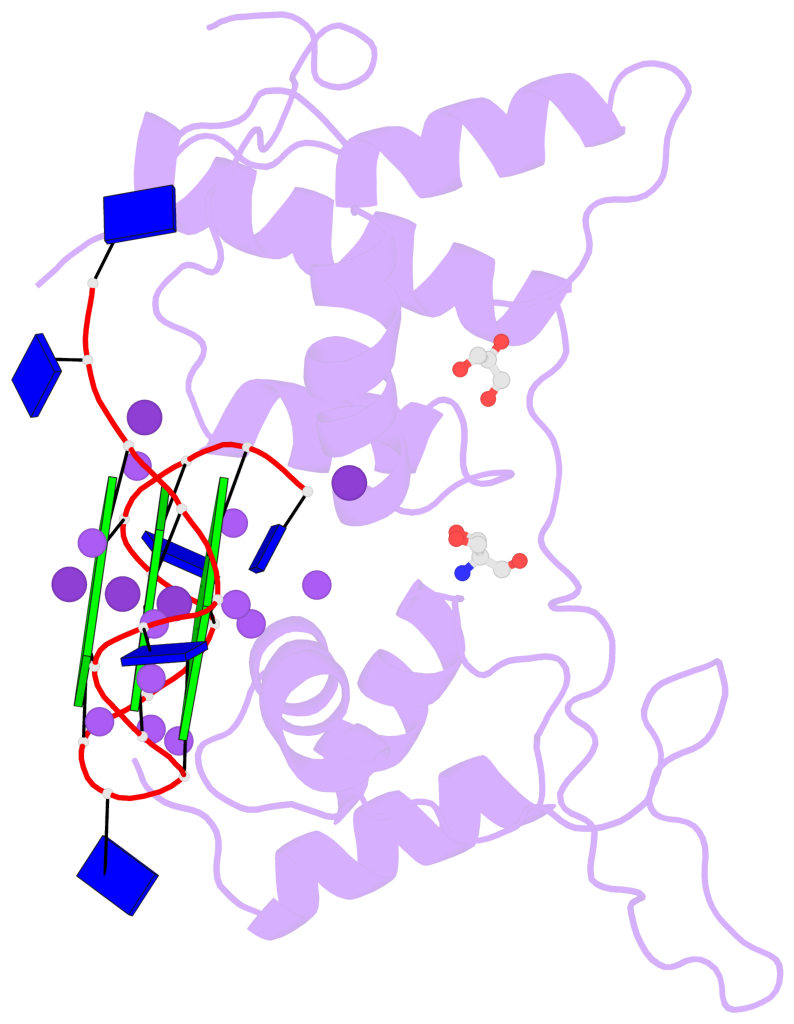
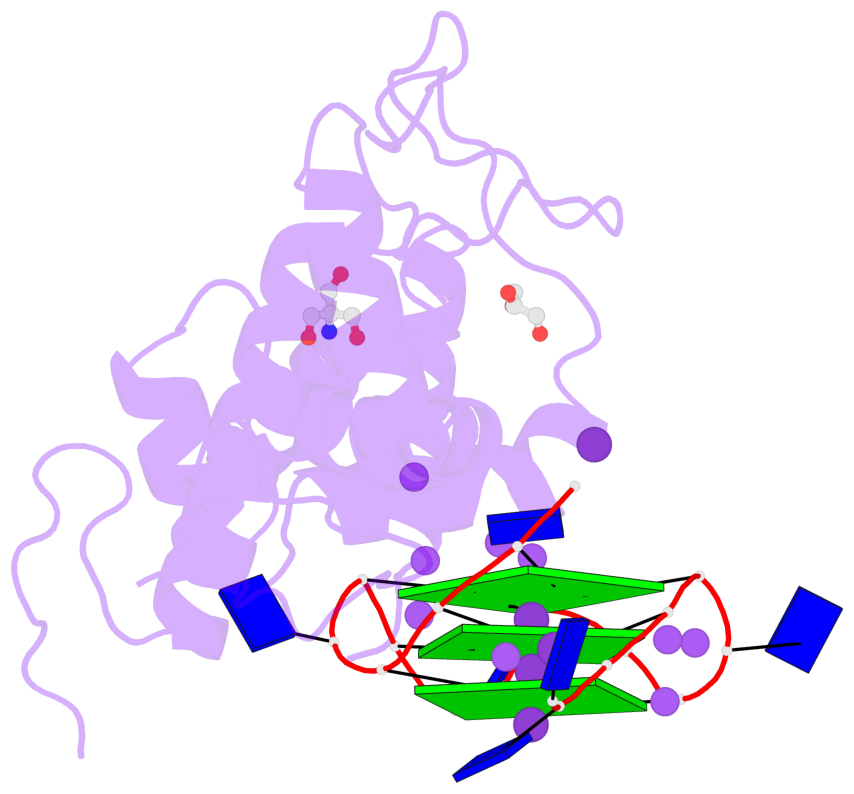
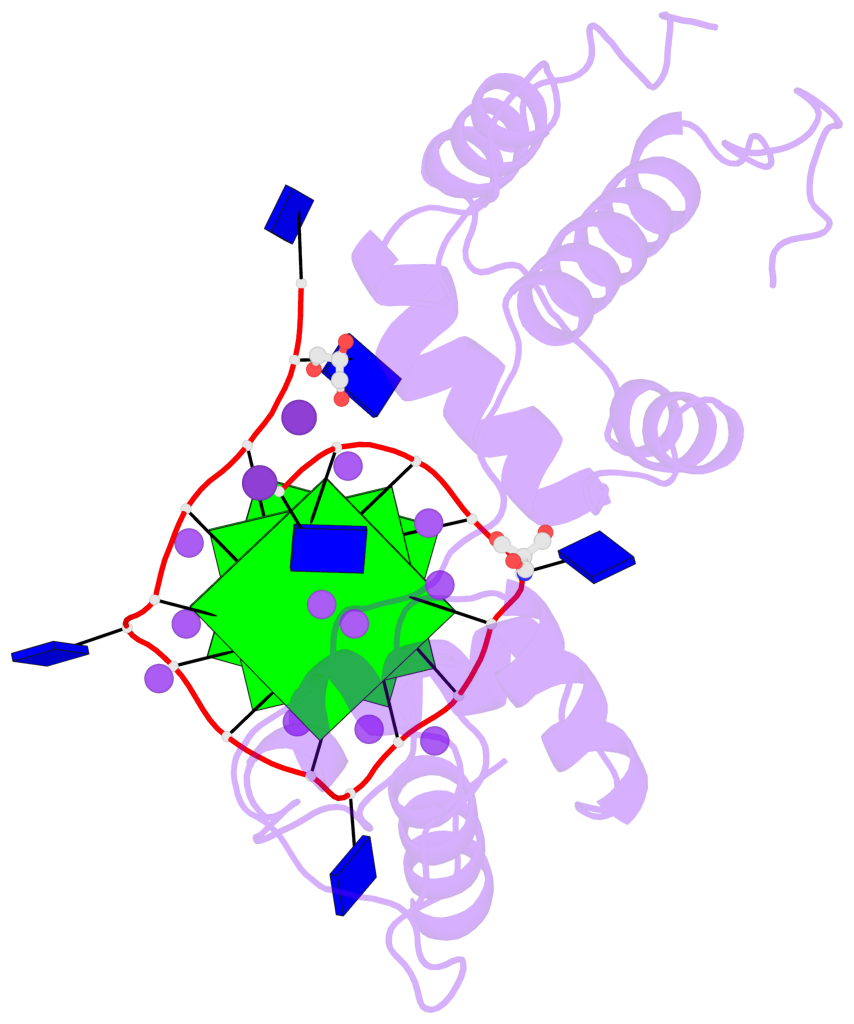
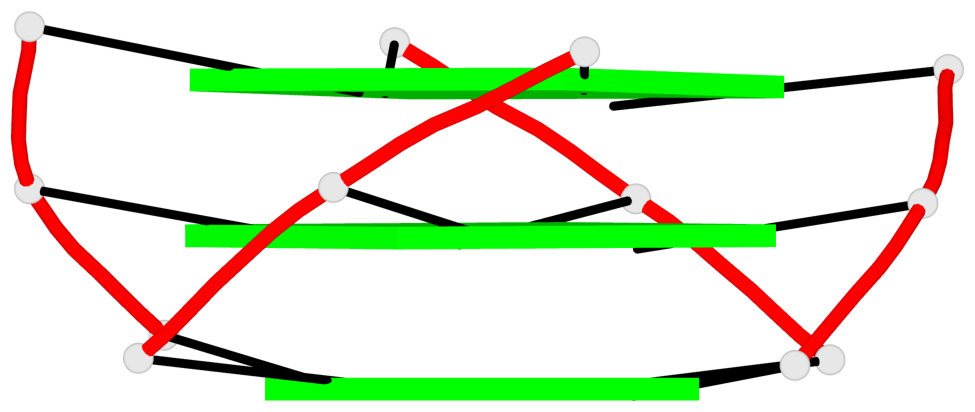
Base-block schematics in six views
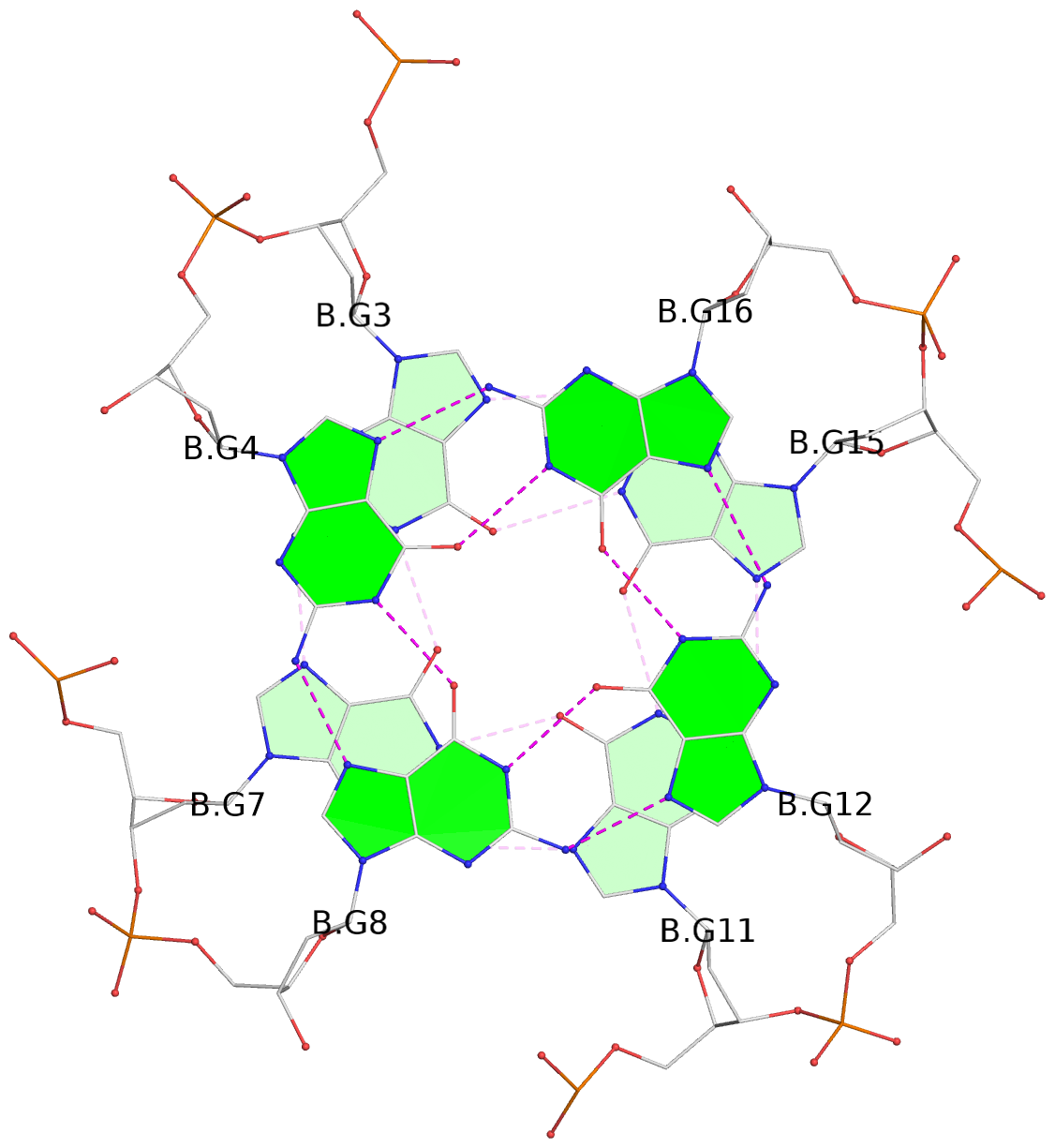
List of 3 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.078 type=planar nts=4 GGGG B.DG3,B.DG7,B.DG11,B.DG15 2 glyco-bond=---- sugar=---- groove=---- planarity=0.155 type=planar nts=4 GGGG B.DG4,B.DG8,B.DG12,B.DG16 3 glyco-bond=---- sugar=---- groove=---- planarity=0.225 type=bowl nts=4 GGGG B.DG5,B.DG9,B.DG13,B.DG17
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.