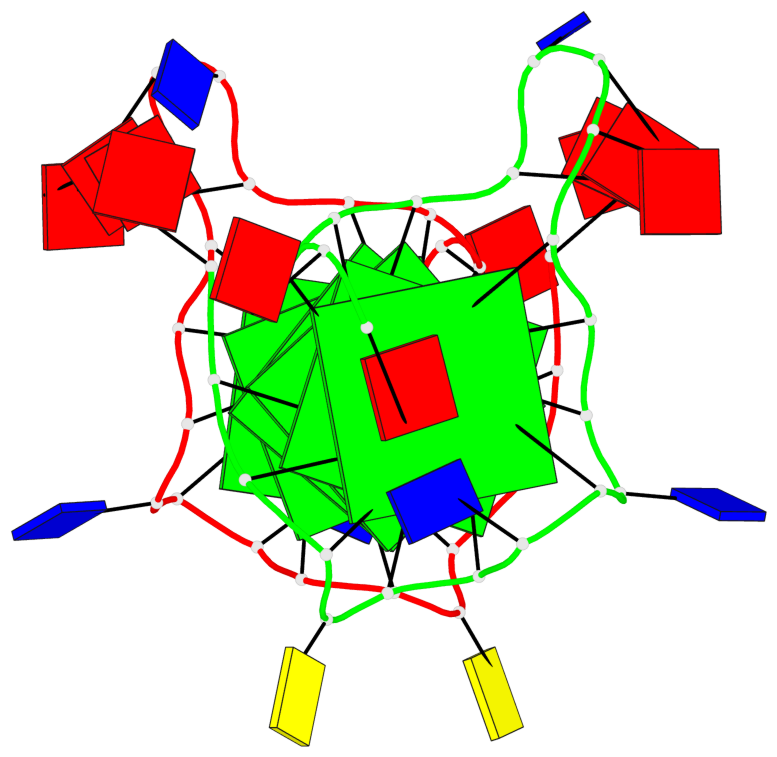
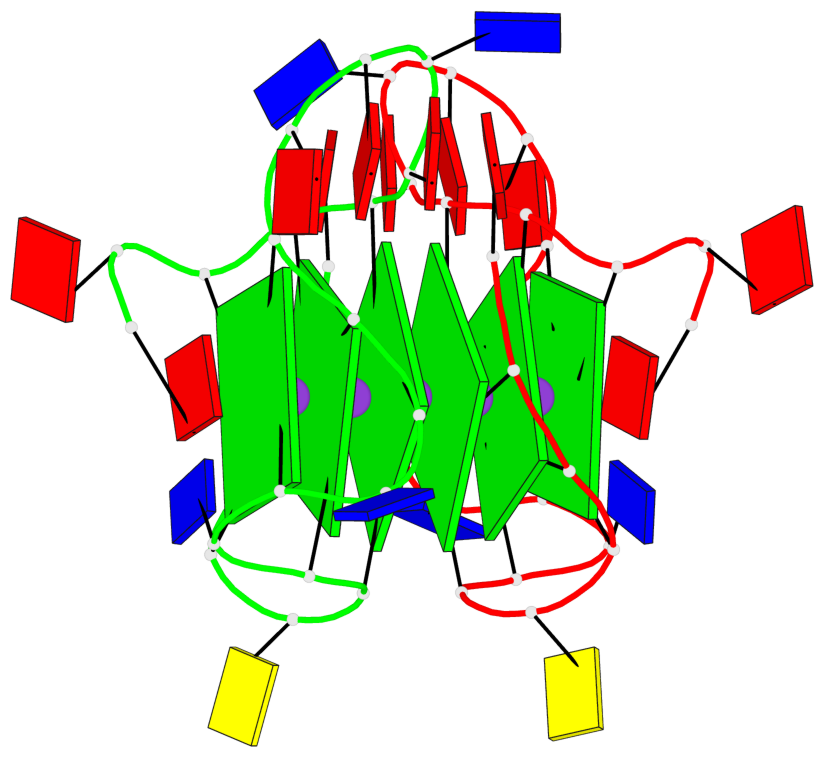
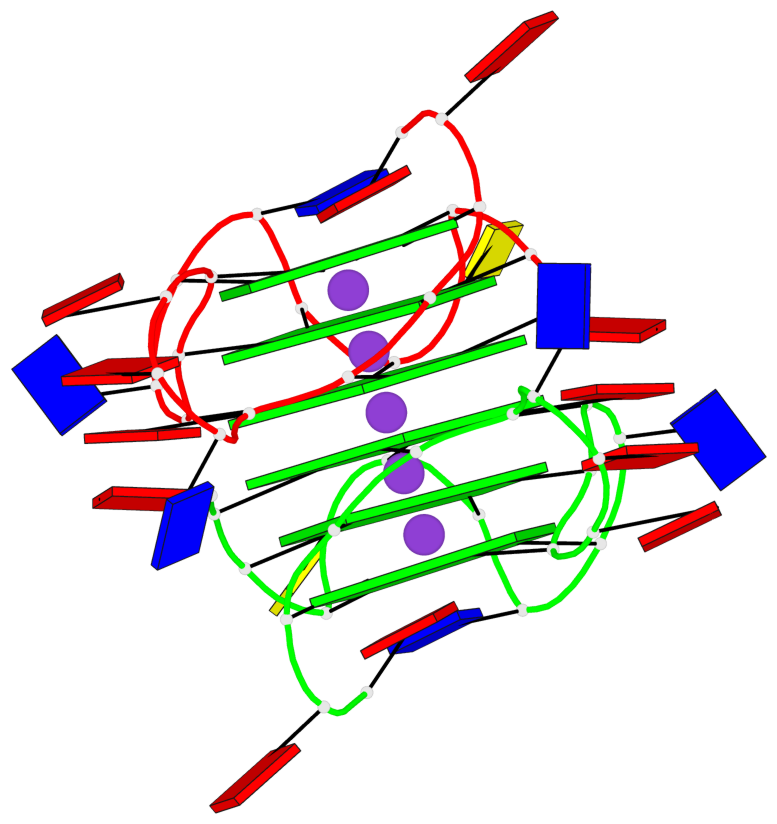
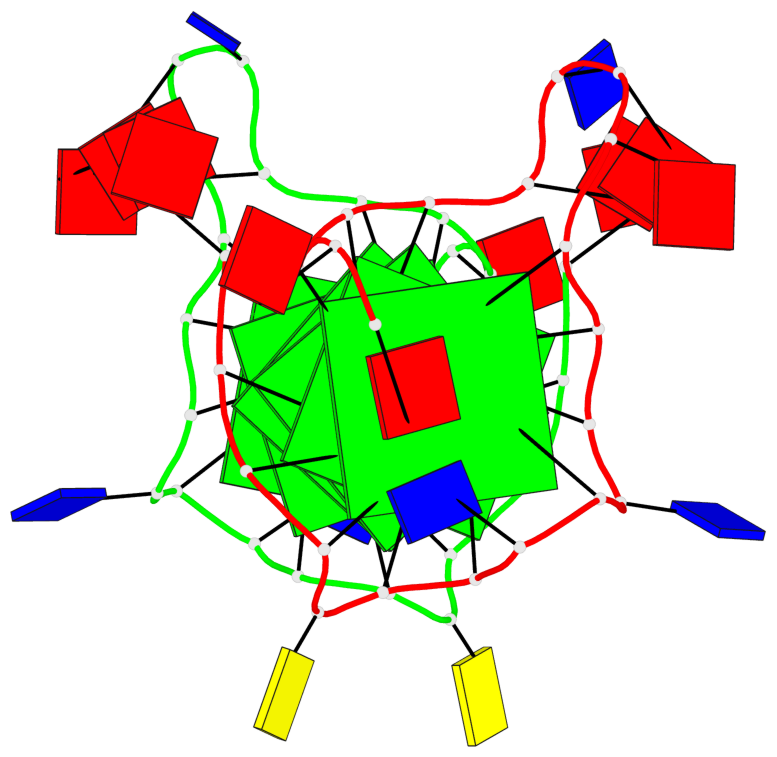
Detailed DSSR results for the G-quadruplex: PDB entry 6n65
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 6n65
- Class
- DNA
- Method
- X-ray (1.6 Å)
- Summary
- Kras g-quadruplex g16t mutant.
- Reference
- Ou A, Schmidberger JW, Wilson KA, Evans CW, Hargreaves JA, Grigg M, O'Mara ML, Iyer KS, Bond CS, Smith NM (2020): "High resolution crystal structure of a KRAS promoter G-quadruplex reveals a dimer with extensive poly-A pi-stacking interactions for small-molecule recognition." Nucleic Acids Res., 48, 5766-5776. doi: 10.1093/nar/gkaa262.
- Abstract
- Aberrant KRAS signaling is a driver of many cancers and yet remains an elusive target for drug therapy. The nuclease hypersensitive element of the KRAS promoter has been reported to form secondary DNA structures called G-quadruplexes (G4s) which may play important roles in regulating KRAS expression, and has spurred interest in structural elucidation studies of the KRAS G-quadruplexes. Here, we report the first high-resolution crystal structure (1.6 Å) of a KRAS G-quadruplex as a 5'-head-to-head dimer with extensive poly-A π-stacking interactions observed across the dimer. Molecular dynamics simulations confirmed that the poly-A π-stacking interactions are also maintained in the G4 monomers. Docking and molecular dynamics simulations with two G4 ligands that display high stabilization of the KRAS G4 indicated the poly-A loop was a binding site for these ligands in addition to the 5'-G-tetrad. Given sequence and structural variability in the loop regions provide the opportunity for small-molecule targeting of specific G4s, we envisage this high-resolution crystal structure for the KRAS G-quadruplex will aid in the rational design of ligands to selectively target KRAS.
- G4 notes
- 6 G-tetrads, 1 G4 helix, 2 G4 stems, 1 G4 coaxial stack, 3(-P-P-P), parallel(4+0), UUUU, coaxial interfaces: 5'/5'
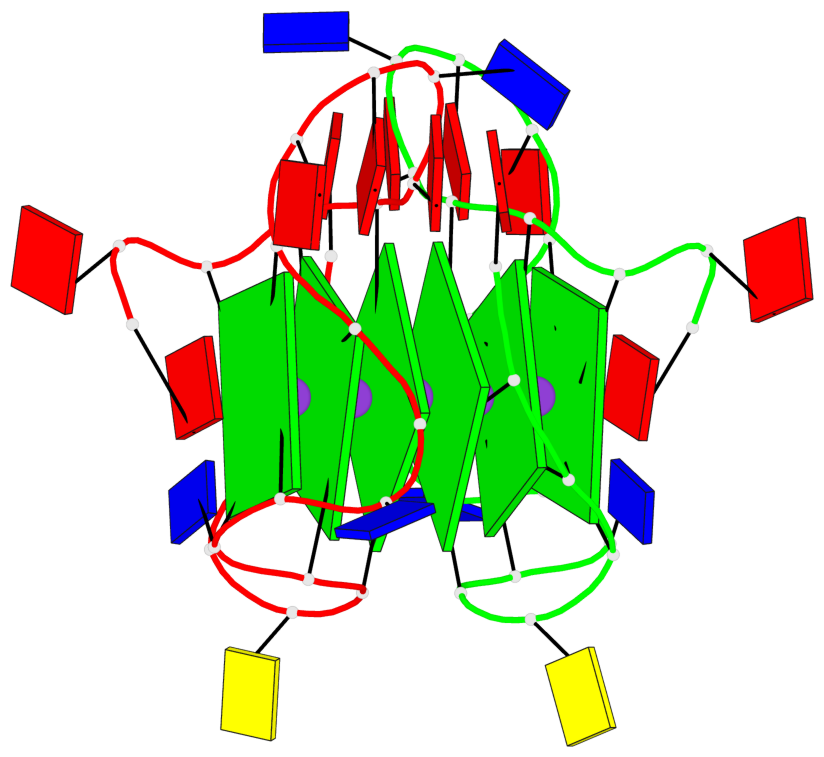
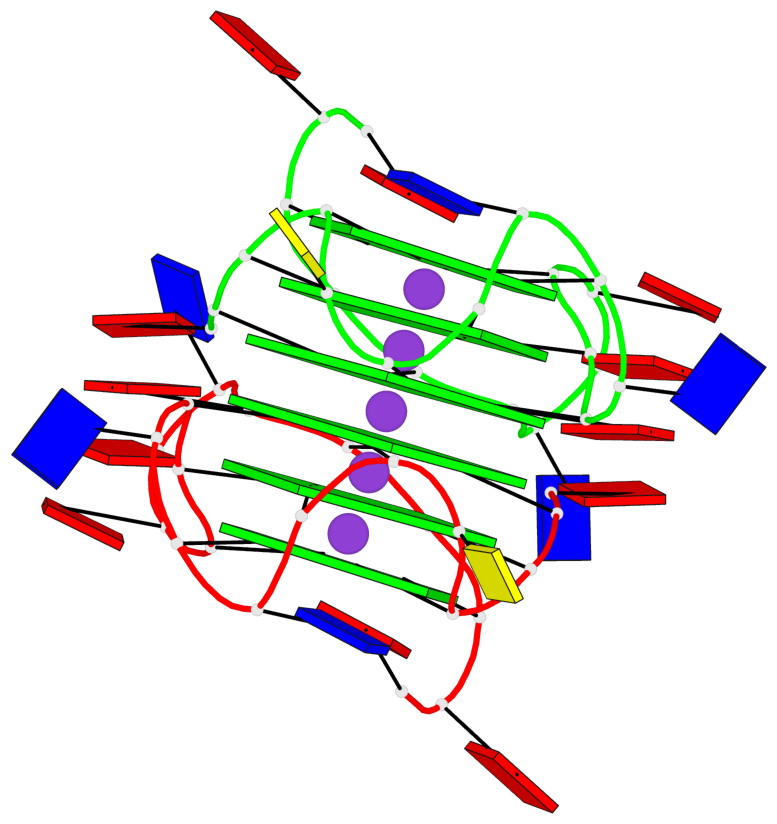
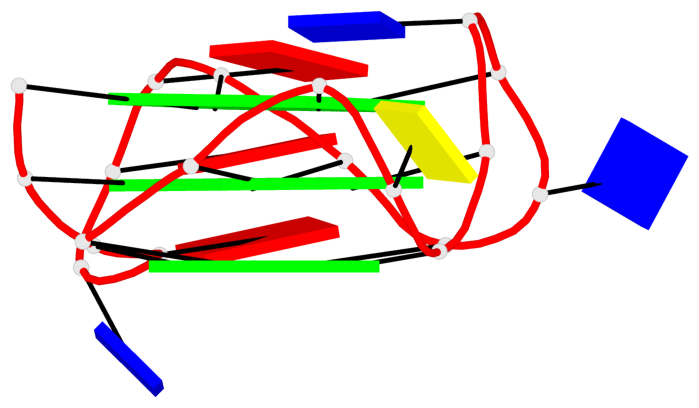
Base-block schematics in six views
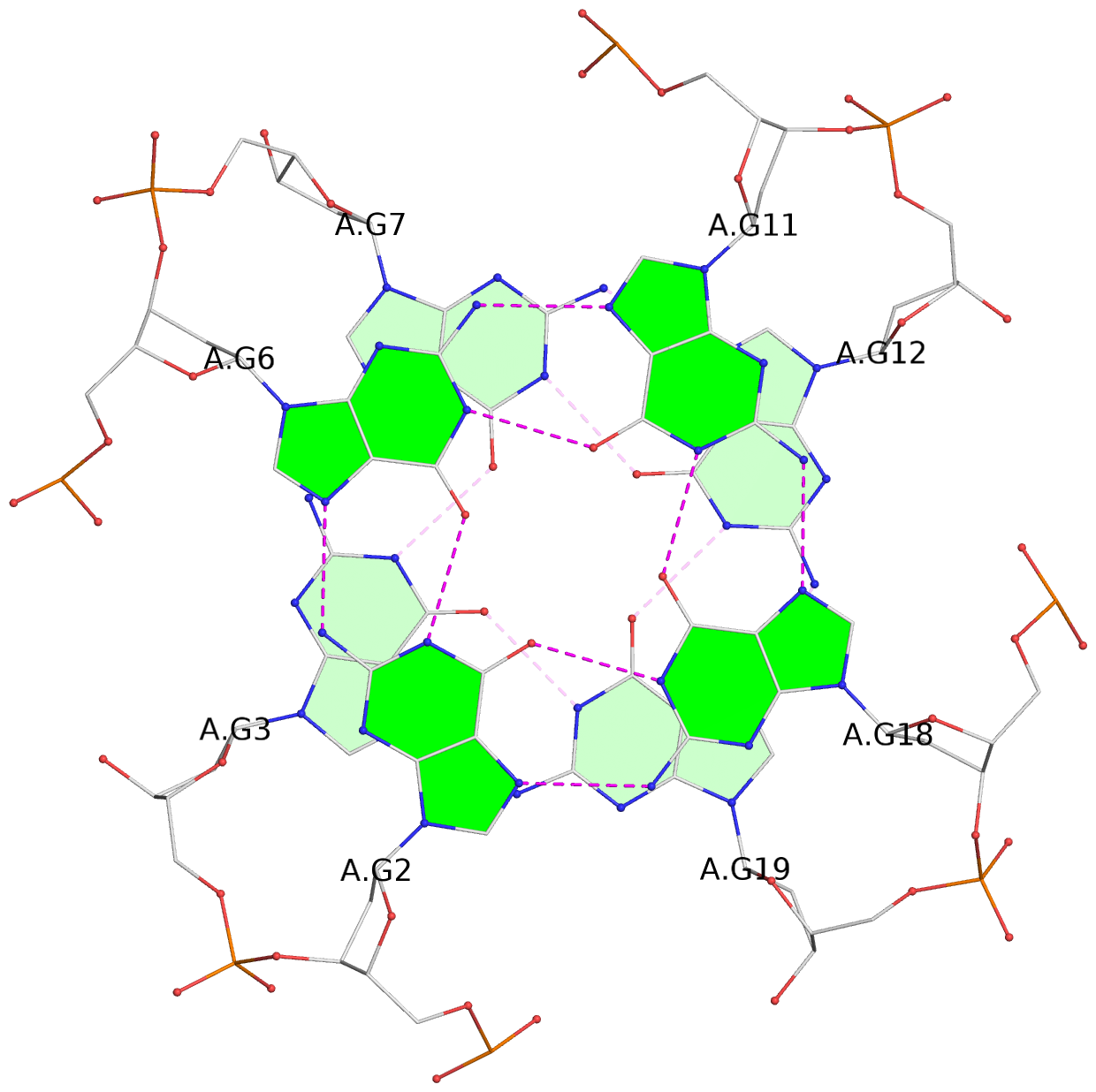
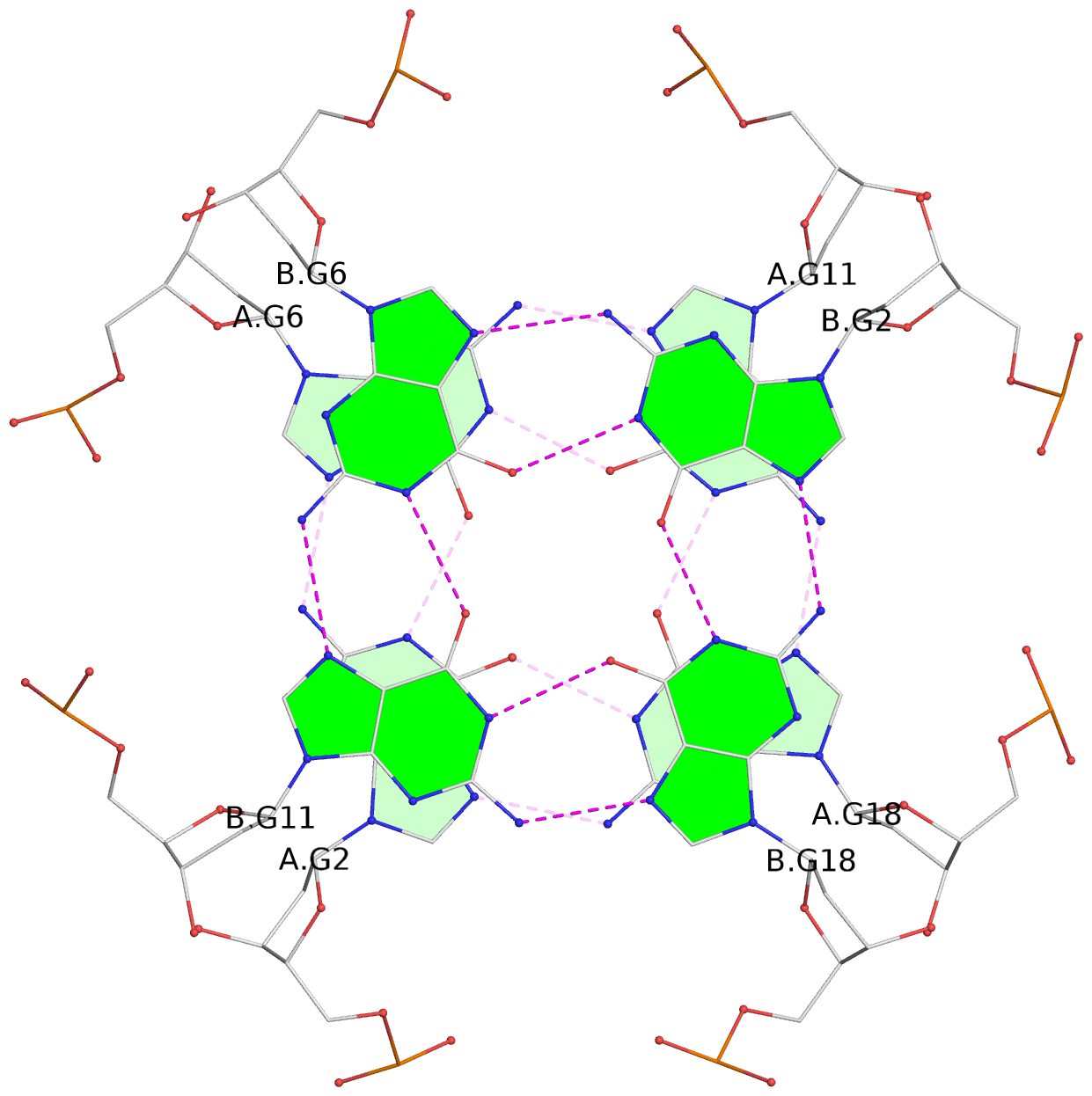
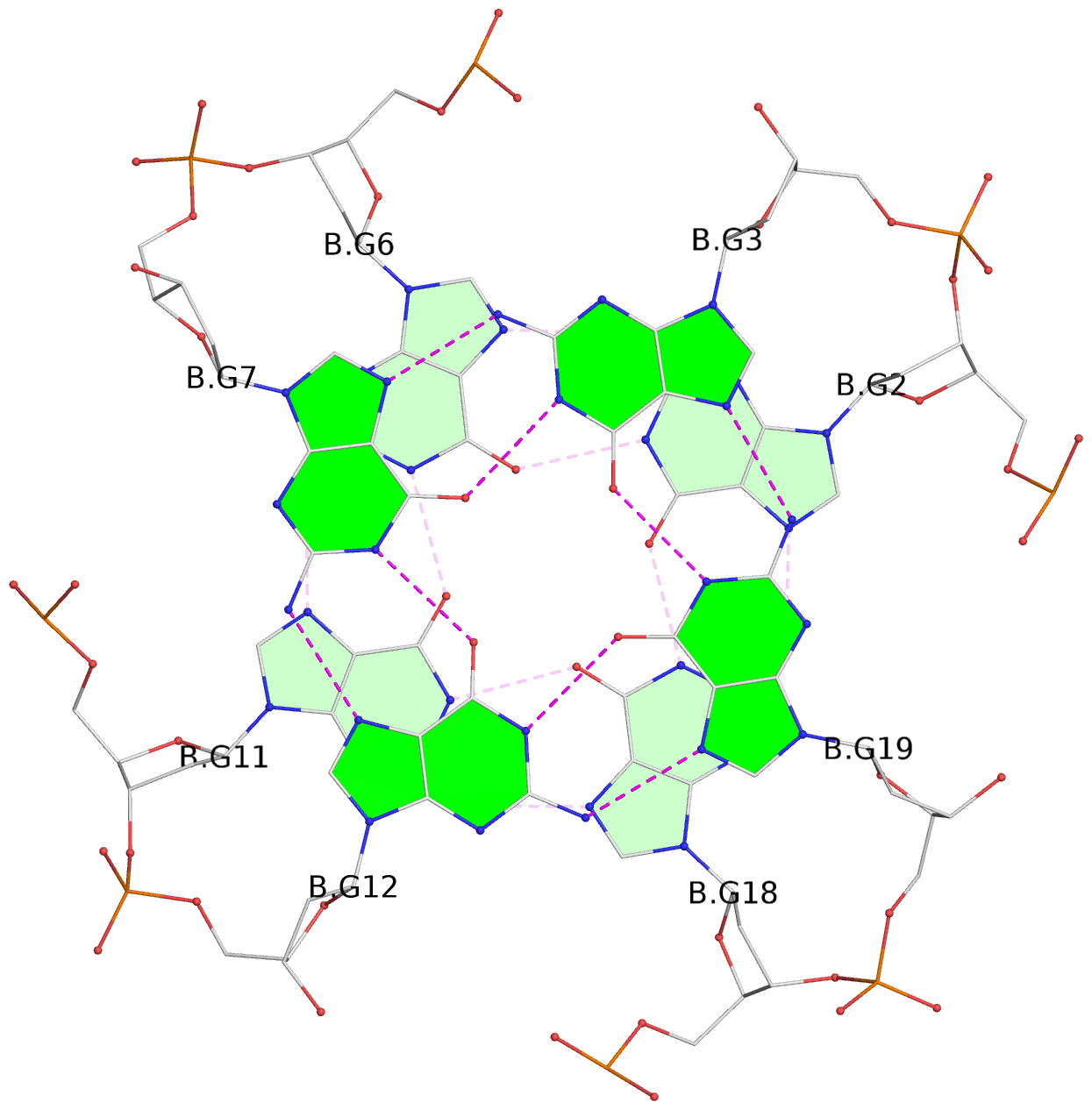
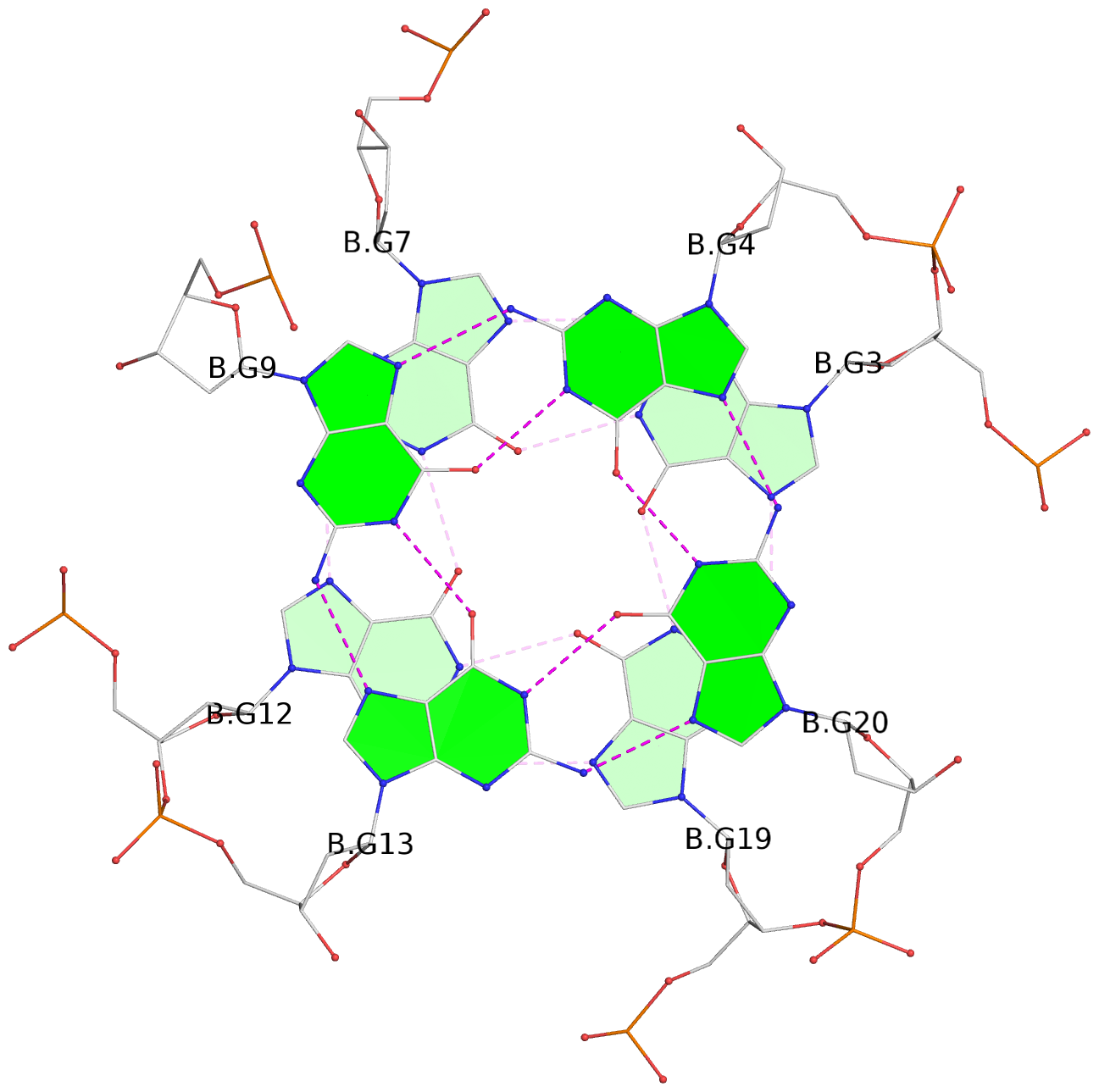
List of 6 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.040 type=planar nts=4 GGGG A.DG2,A.DG6,A.DG11,A.DG18 2 glyco-bond=---- sugar=-.-- groove=---- planarity=0.127 type=planar nts=4 GGGG A.DG3,A.DG7,A.DG12,A.DG19 3 glyco-bond=---- sugar=---- groove=---- planarity=0.206 type=bowl nts=4 GGGG A.DG4,A.DG9,A.DG13,A.DG20 4 glyco-bond=---- sugar=---- groove=---- planarity=0.041 type=planar nts=4 GGGG B.DG2,B.DG6,B.DG11,B.DG18 5 glyco-bond=---- sugar=-.-- groove=---- planarity=0.121 type=planar nts=4 GGGG B.DG3,B.DG7,B.DG12,B.DG19 6 glyco-bond=---- sugar=---- groove=---- planarity=0.205 type=bowl nts=4 GGGG B.DG4,B.DG9,B.DG13,B.DG20
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 6 G-tetrad layers, inter-molecular, with 2 stems
List of 2 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 3 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 3(-P-P-P), parallel(4+0)
Stem#2, 3 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 3(-P-P-P), parallel(4+0)
List of 1 G4 coaxial stack
1 G4 helix#1 contains 2 G4 stems: [#1,#2] [5'/5']