Detailed DSSR results for the G-quadruplex: PDB entry 6pnk
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 6pnk
- Class
- DNA
- Method
- X-ray (2.39 Å)
- Summary
- Crystal structure of the g-quadruplex formed by (gggtt)3ggg in complex with n-methylmesoporphryin ix
- Reference
- Lin LY, McCarthy S, Powell BM, Manurung Y, Xiang IM, Dean WL, Chaires B, Yatsunyk LA (2020): "Biophysical and X-ray structural studies of the (GGGTT)3GGG G-quadruplex in complex with N-methyl mesoporphyrin IX." Plos One, 15, e0241513. doi: 10.1371/journal.pone.0241513.
- Abstract
- The G-quadruplex (GQ) is a well-studied non-canonical DNA structure formed by G-rich sequences found at telomeres and gene promoters. Biological studies suggest that GQs may play roles in regulating gene expression, DNA replication, and DNA repair. Small molecule ligands were shown to alter GQ structure and stability and thereby serve as novel therapies, particularly against cancer. In this work, we investigate the interaction of a G-rich sequence, 5'-GGGTTGGGTTGGGTTGGG-3' (T1), with a water-soluble porphyrin, N-methyl mesoporphyrin IX (NMM) via biophysical and X-ray crystallographic studies. UV-vis and fluorescence titrations, as well as a Job plot, revealed a 1:1 binding stoichiometry with an impressively tight binding constant of 30-50 μM-1 and ΔG298 of -10.3 kcal/mol. Eight extended variants of T1 (named T2 -T9) were fully characterized and T7 was identified as a suitable candidate for crystallographic studies. We solved the crystal structures of the T1- and T7-NMM complexes at 2.39 and 2.34 Å resolution, respectively. Both complexes form a 5'-5' dimer of parallel GQs capped by NMM at the 3' G-quartet, supporting the 1:1 binding stoichiometry. Our work provides invaluable details about GQ-ligand binding interactions and informs the design of novel anticancer drugs that selectively recognize specific GQs and modulate their stability for therapeutic purposes.
- G4 notes
- 6 G-tetrads, 2 G4 helices, 2 G4 stems, 3(-P-P), parallel(4+0), UUUU; 3(-P-P-P), parallel(4+0), UUUU
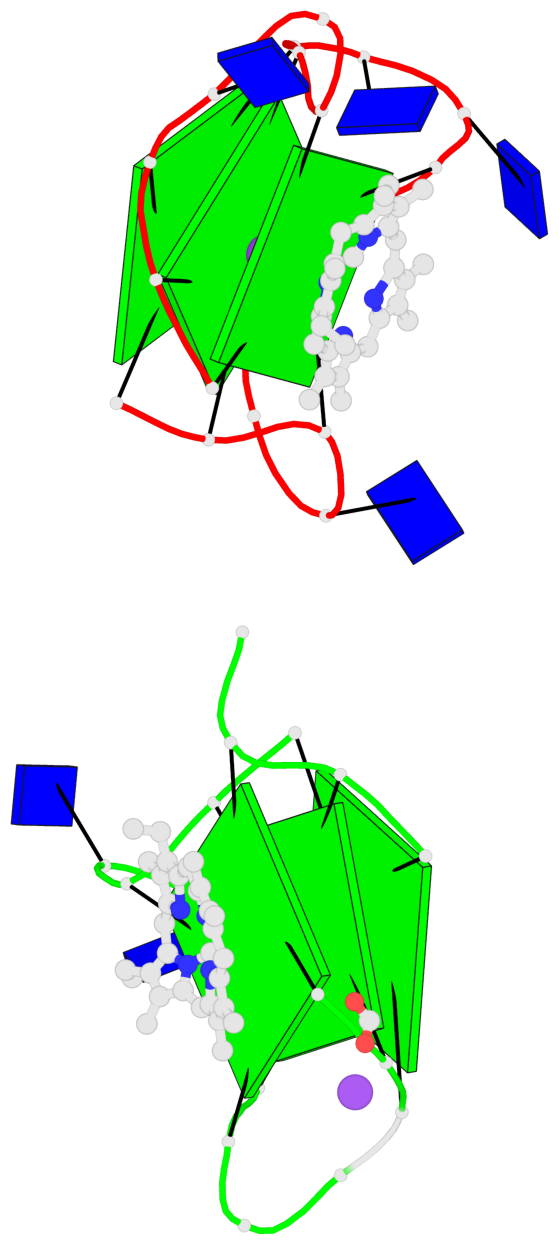
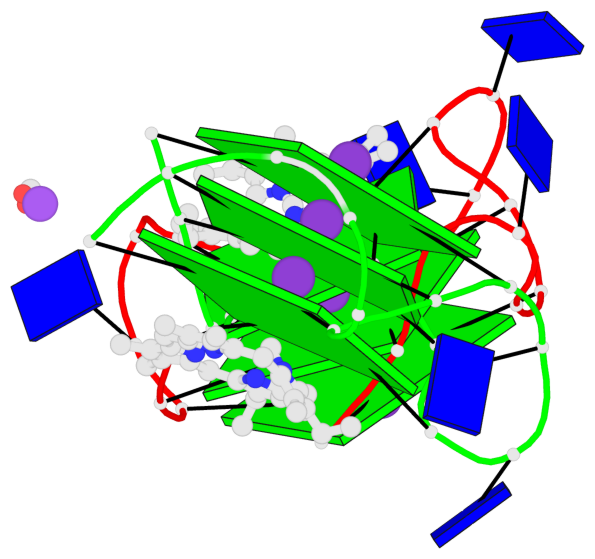
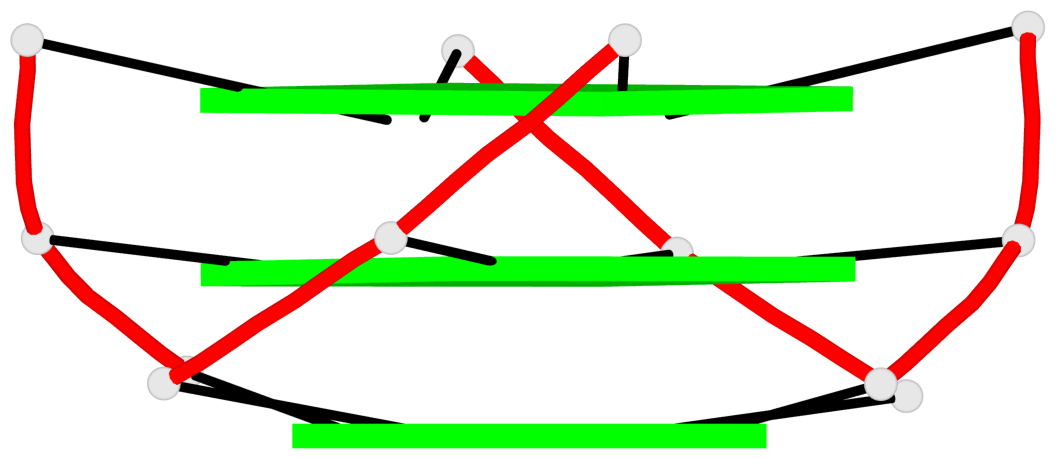
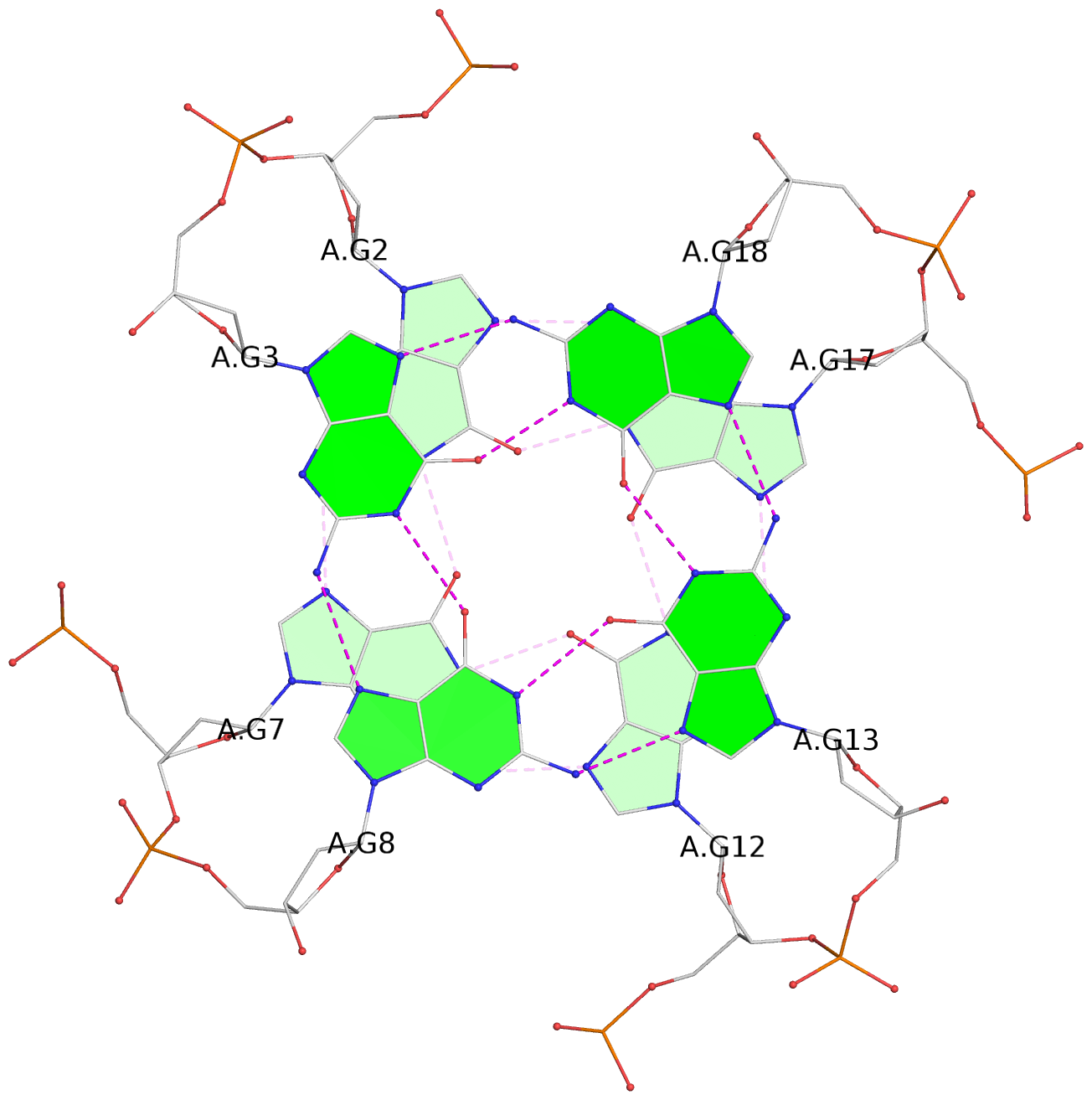
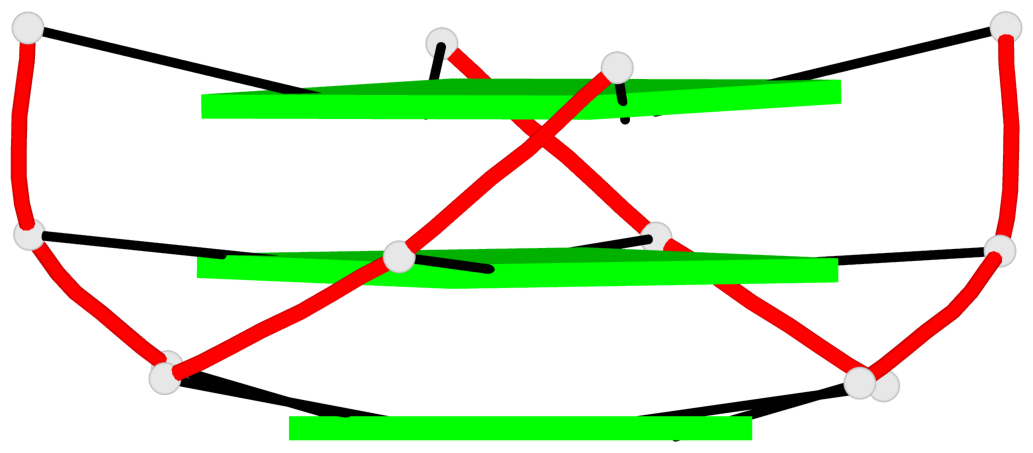
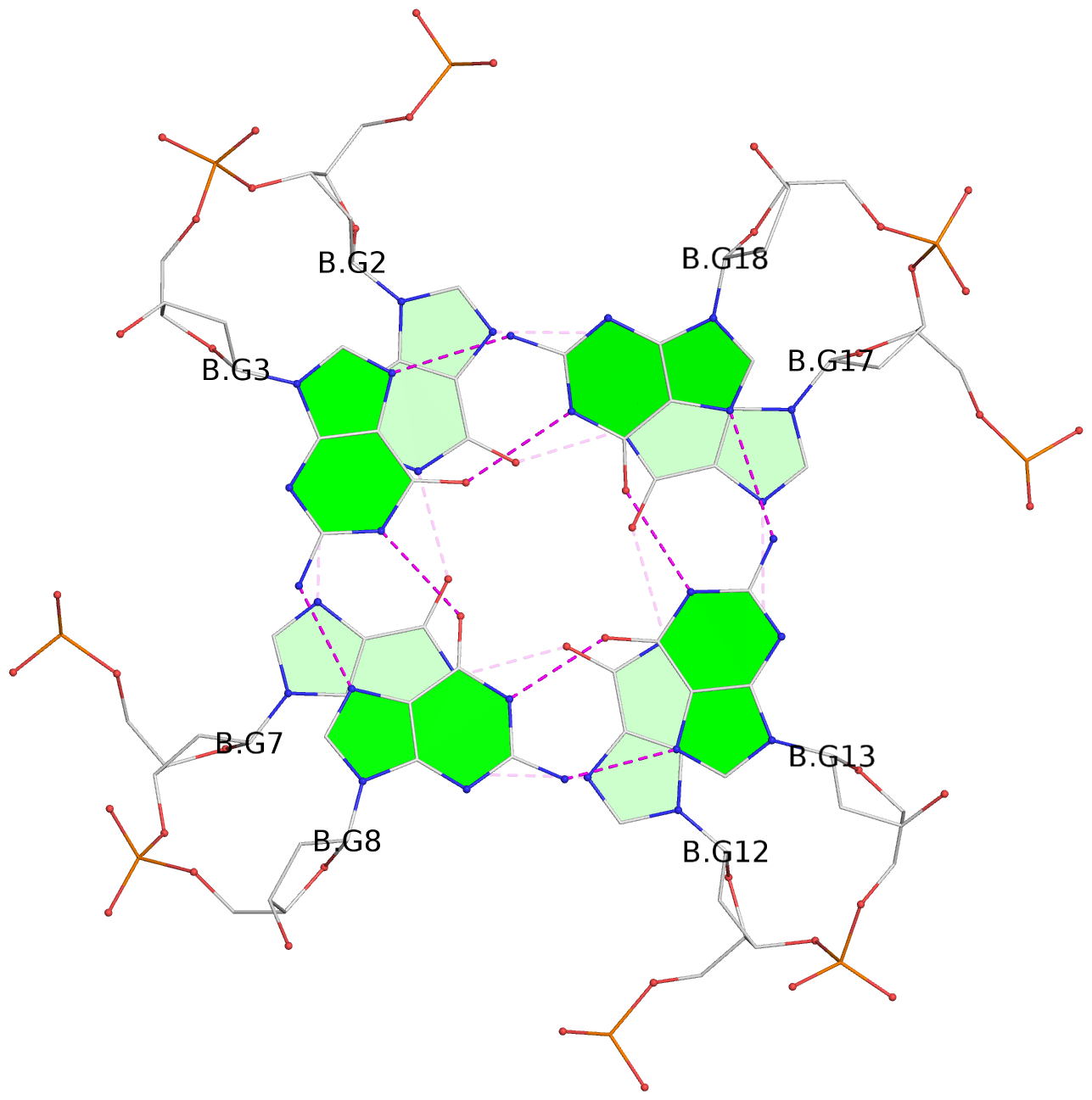
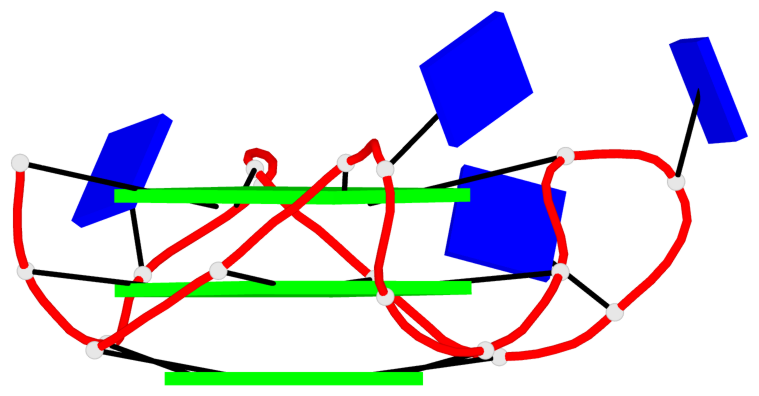
Base-block schematics in six views
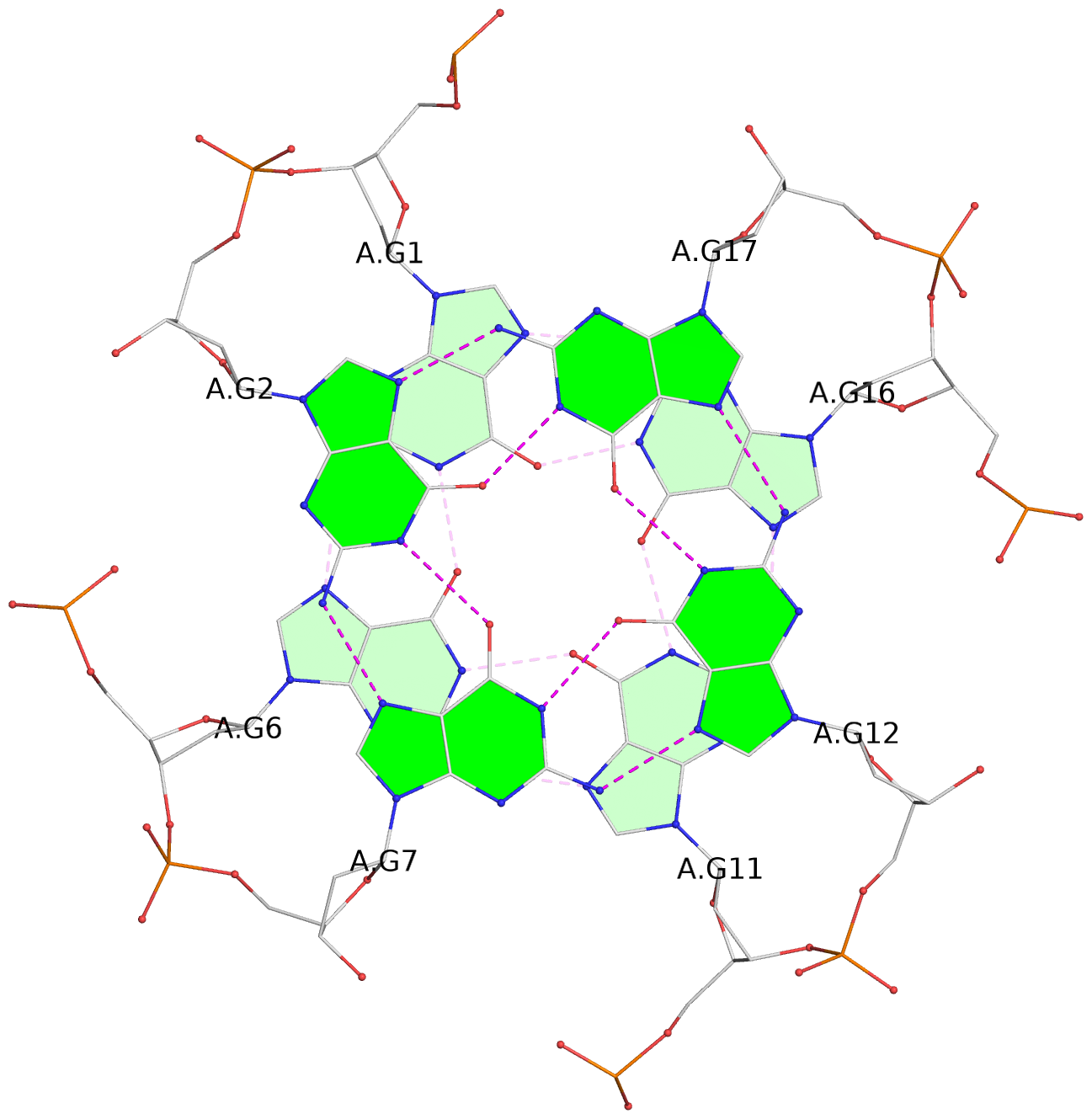
List of 6 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.074 type=planar nts=4 GGGG A.DG1,A.DG6,A.DG11,A.DG16 2 glyco-bond=---- sugar=---- groove=---- planarity=0.095 type=planar nts=4 GGGG A.DG2,A.DG7,A.DG12,A.DG17 3 glyco-bond=---- sugar=---- groove=---- planarity=0.295 type=bowl nts=4 GGGG A.DG3,A.DG8,A.DG13,A.DG18 4 glyco-bond=---- sugar=---- groove=---- planarity=0.061 type=planar nts=4 GGGG B.DG1,B.DG6,B.DG11,B.DG16 5 glyco-bond=---- sugar=---- groove=---- planarity=0.060 type=planar nts=4 GGGG B.DG2,B.DG7,B.DG12,B.DG17 6 glyco-bond=---- sugar=---- groove=---- planarity=0.318 type=bowl nts=4 GGGG B.DG3,B.DG8,B.DG13,B.DG18
List of 2 G4-helices
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
Helix#2, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 2 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.