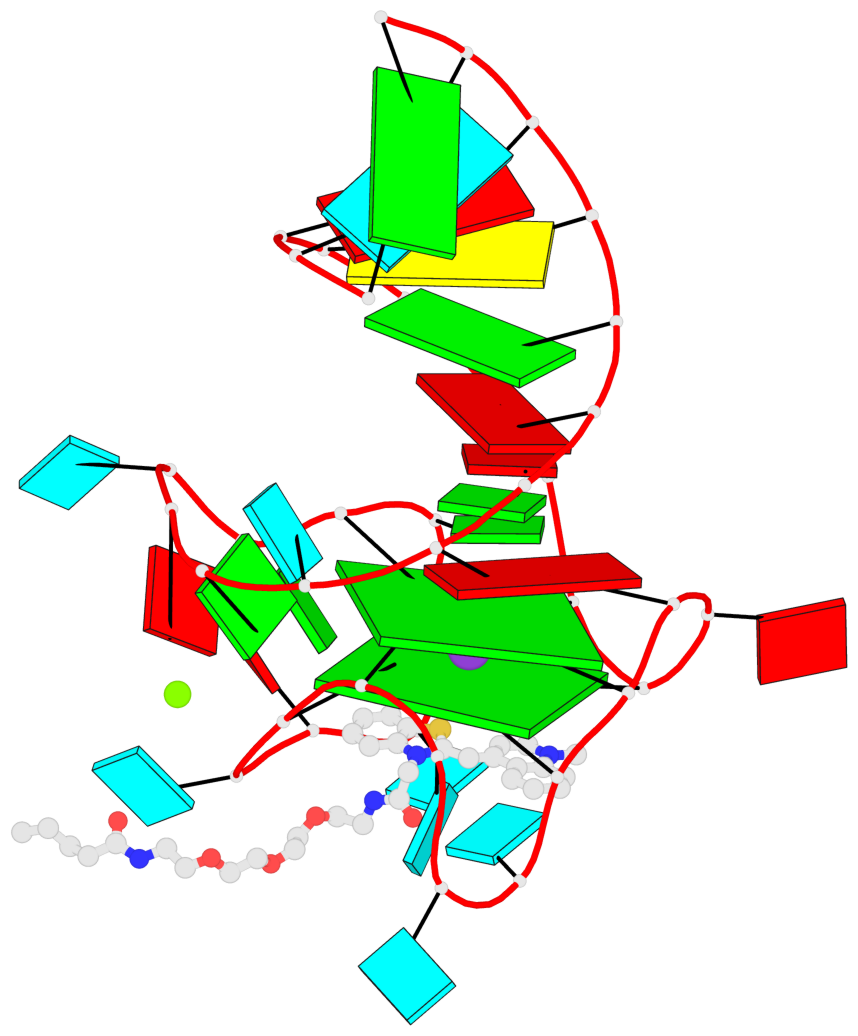
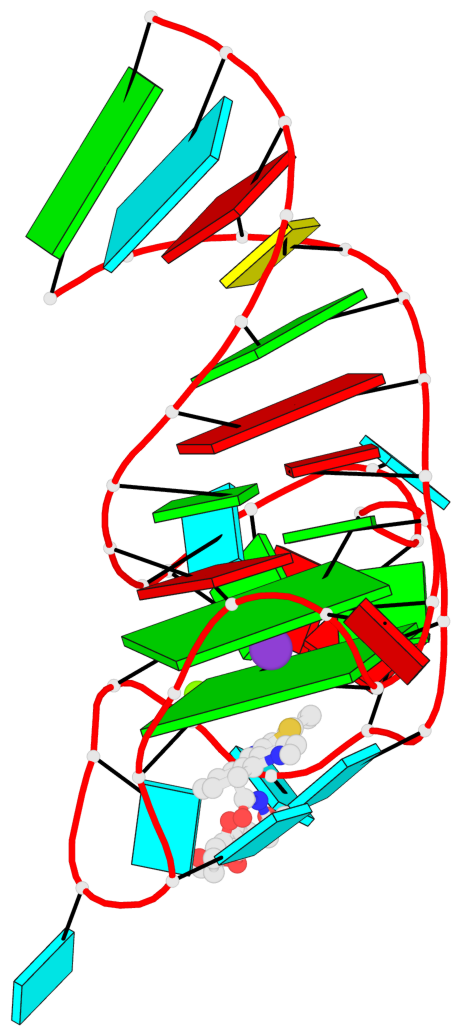
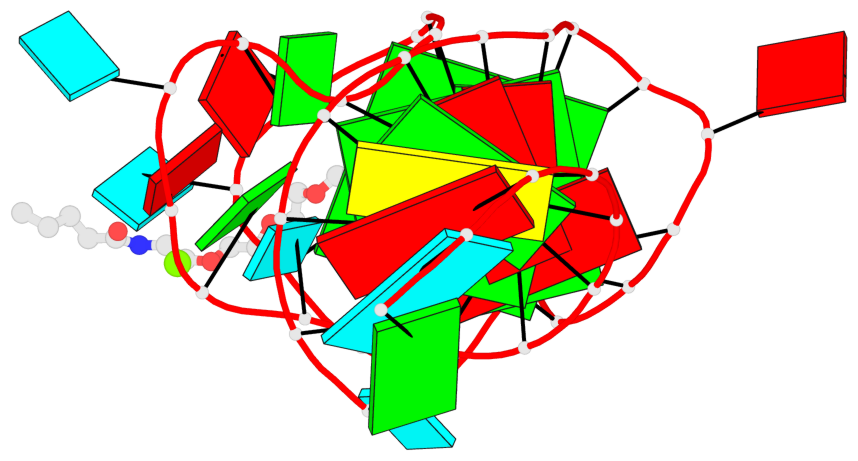
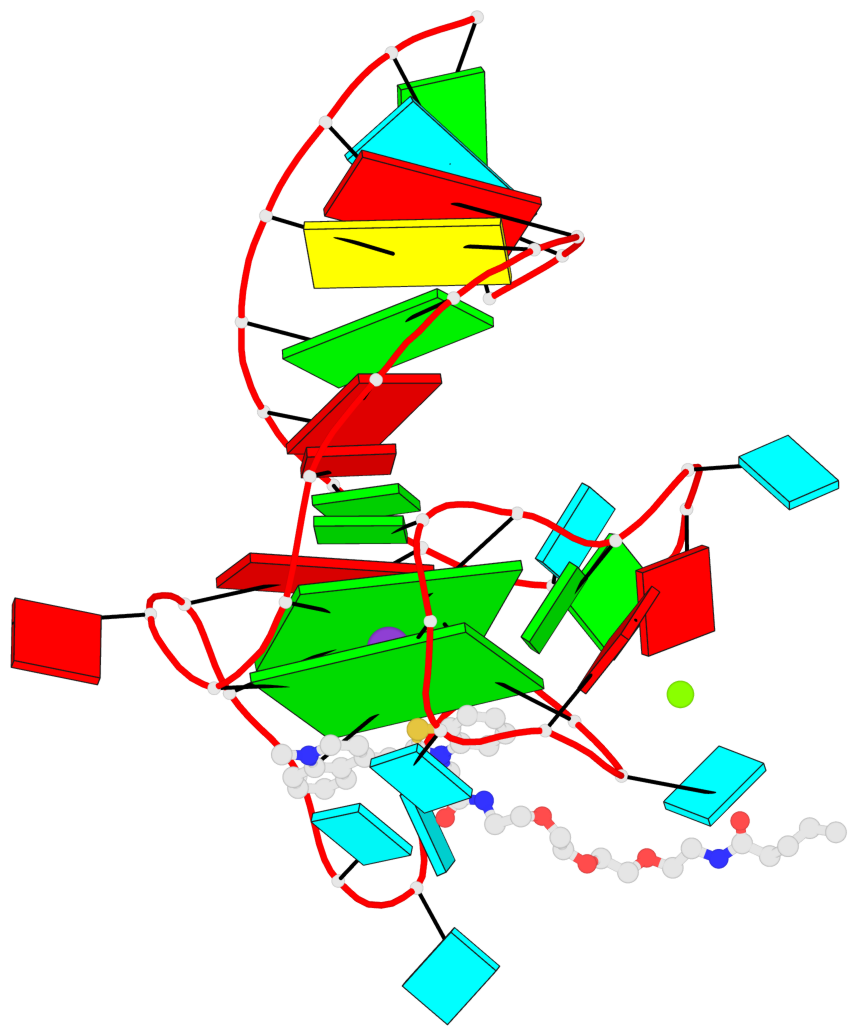
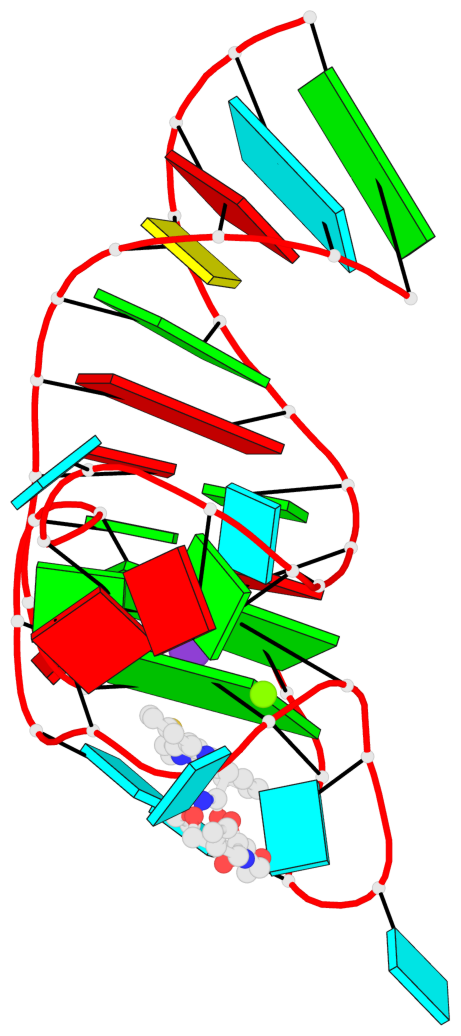
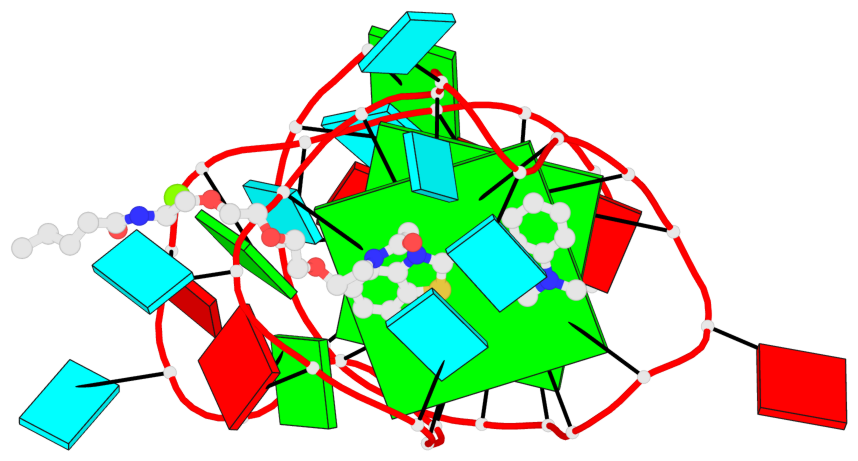
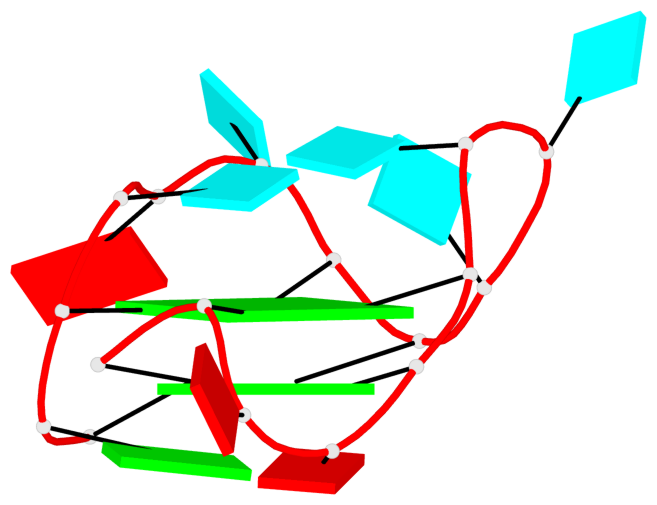
Detailed DSSR results for the G-quadruplex: PDB entry 6pq7
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 6pq7
- Class
- RNA
- Method
- X-ray (3.0 Å)
- Summary
- Structure of the imango-iii fluorescent aptamer at room temperature.
- Reference
- Trachman III RJ, Stagno JR, Conrad C, Jones CP, Fischer P, Meents A, Wang YX, Ferre-D'Amare AR (2019): "Co-crystal structure of the iMango-III fluorescent RNA aptamer using an X-ray free-electron laser." Acta Crystallogr.,Sect.F, 75, 547-551. doi: 10.1107/S2053230X19010136.
- Abstract
- Turn-on aptamers are in vitro-selected RNAs that bind to conditionally fluorescent small molecules and enhance their fluorescence. Upon binding TO1-biotin, the iMango-III aptamer achieves the largest fluorescence enhancement reported for turn-on aptamers (over 5000-fold). This aptamer was generated by structure-guided engineering and functional reselection of the parental aptamer Mango-III. Structures of both Mango-III and iMango-III have previously been determined by conventional cryocrystallography using synchrotron X-radiation. Using an X-ray free-electron laser (XFEL), the room-temperature iMango-III-TO1-biotin co-crystal structure has now been determined at 3.0 Å resolution. This structural model, which was refined against a data set of ∼1300 diffraction images (each from a single crystal), is largely consistent with the structures determined from single-crystal data sets collected at 100 K. This constitutes a technical benchmark on the way to XFEL pump-probe experiments on fluorescent RNA-small molecule complexes.
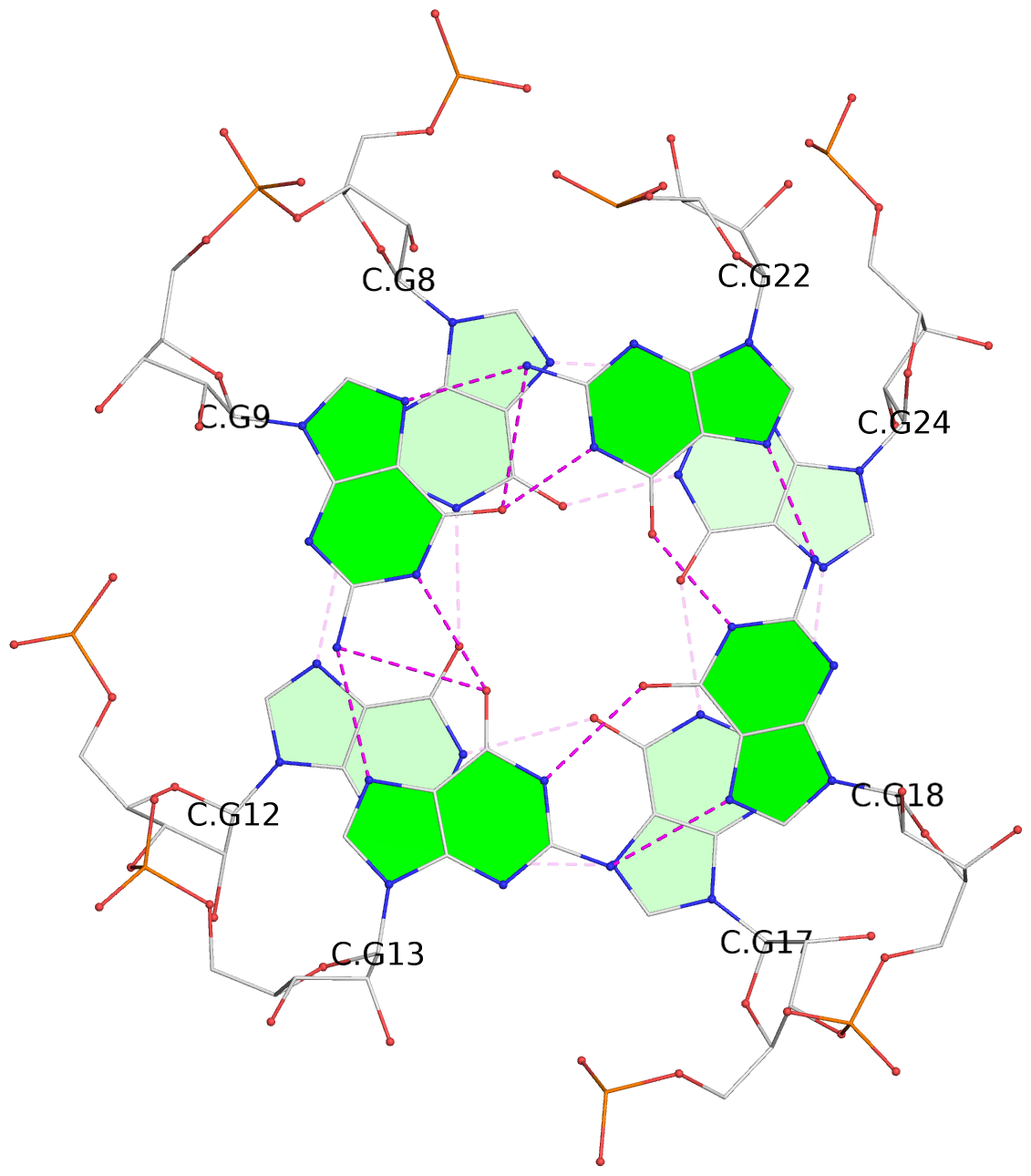
- G4 notes
- 2 G-tetrads, 1 G4 helix, 1 G4 stem, 2(-P-P-Lw), hybrid-2R(3+1), UUUD
Base-block schematics in six views
List of 2 G-tetrads
1 glyco-bond=---s sugar=-33- groove=--wn planarity=0.350 type=other nts=4 GGGG C.G8,C.G12,C.G17,C.G24 2 glyco-bond=---s sugar=-3-3 groove=--wn planarity=0.262 type=other nts=4 GGGG C.G9,C.G13,C.G18,C.G22
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 2 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.