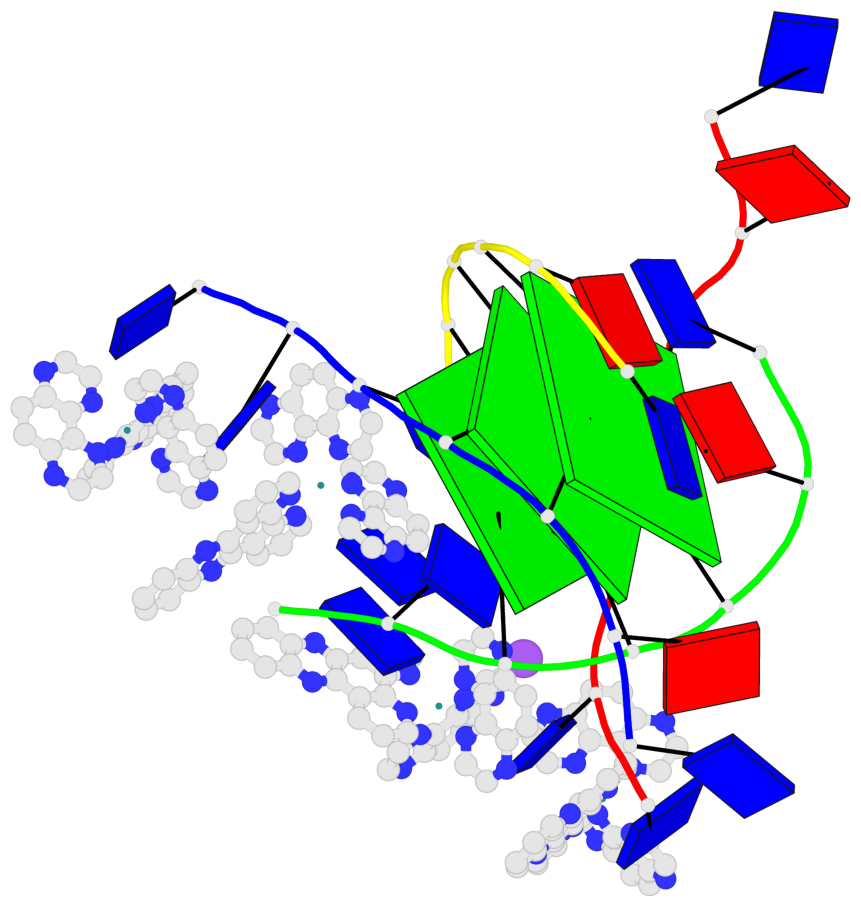
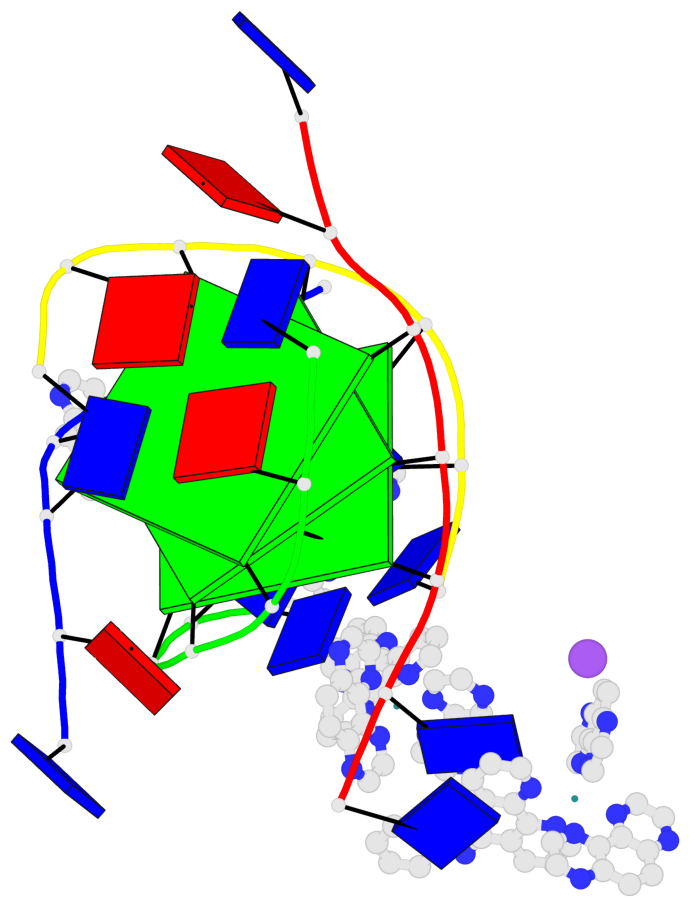
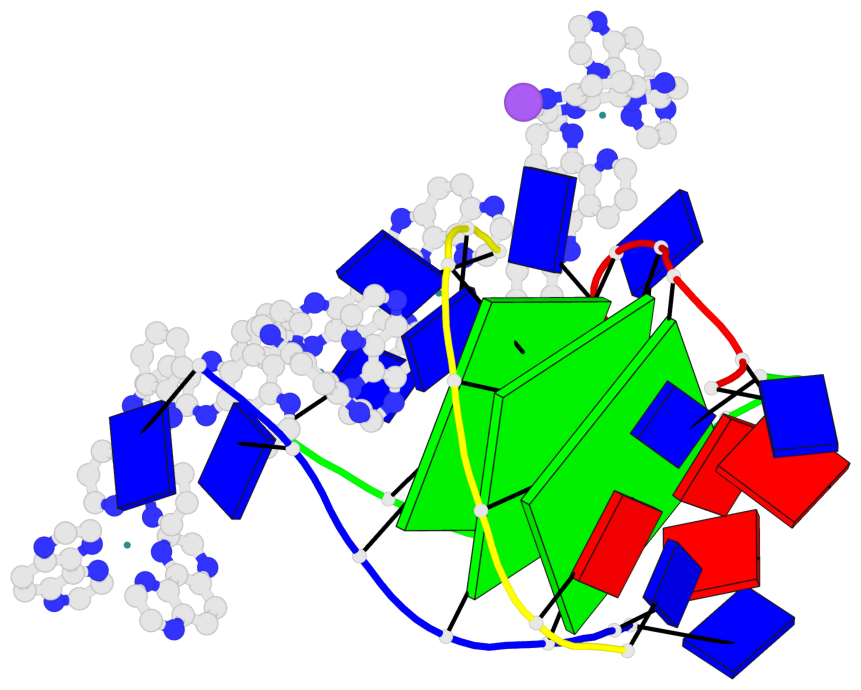
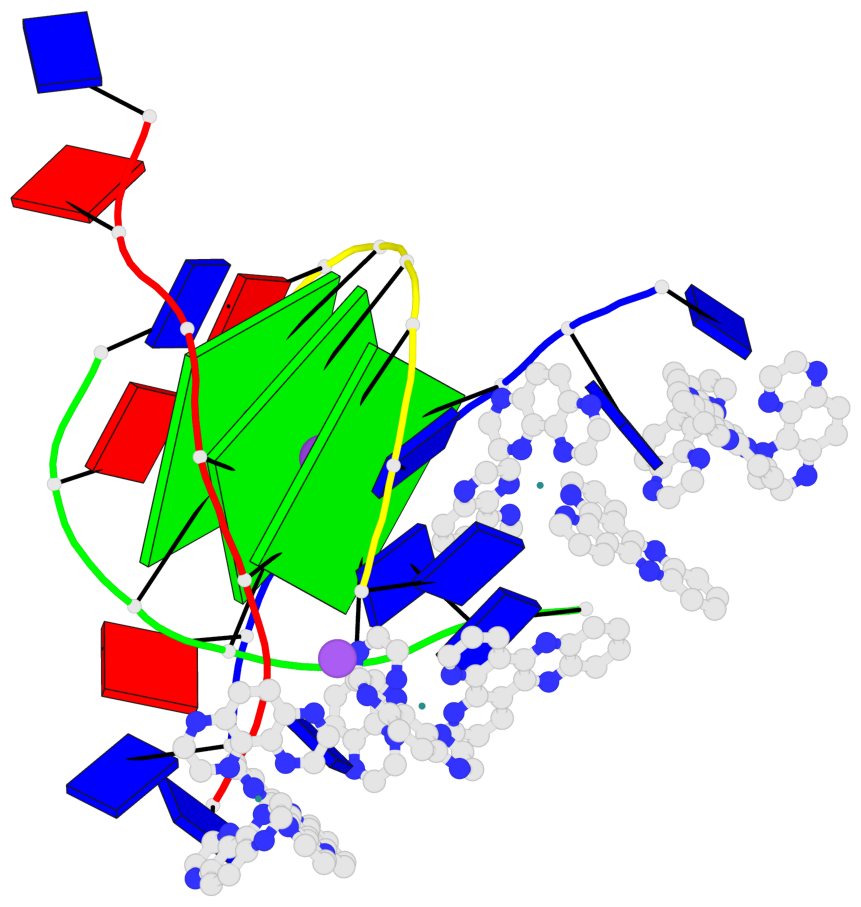
Detailed DSSR results for the G-quadruplex: PDB entry 6rnl
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 6rnl
- Class
- DNA
- Method
- X-ray (1.88 Å)
- Summary
- L-[ru(tap)2(dppz)]2+ bound to the g-quadruplex forming sequence d(tagggtt)
- Reference
- McQuaid K, Hall JP, Baumgaertner L, Cardin DJ, Cardin CJ (2019): "Three thymine/adenine binding modes of the ruthenium complex Lambda-[Ru(TAP)2(dppz)]2+to the G-quadruplex forming sequence d(TAGGGTT) shown by X-ray crystallography." Chem.Commun.(Camb.), 55, 9116-9119. doi: 10.1039/c9cc04316k.
- Abstract
- Λ-[Ru(TAP)2(dppz)]2+ was crystallised with the G-quadruplex-forming heptamer d(TAGGGTT). Surprisingly, even though there are four unique binding sites, the complex is not in contact with any G-quartet surface. Two complexes stabilise cavities formed from terminal T·A and T·T mismatched pairs. A third shows kinking by a TAP ligand between T·T linkages, while the fourth shows sandwiching of a dppz ligand between a T·A/T·A quadruplex and a T·T mismatch, stabilised by an additional T·A base pair stacking interaction on a TAP surface. Overall, the structure shows an unexpected affinity for thymine, and suggests models for G-quadruplex loop binding.
- G4 notes
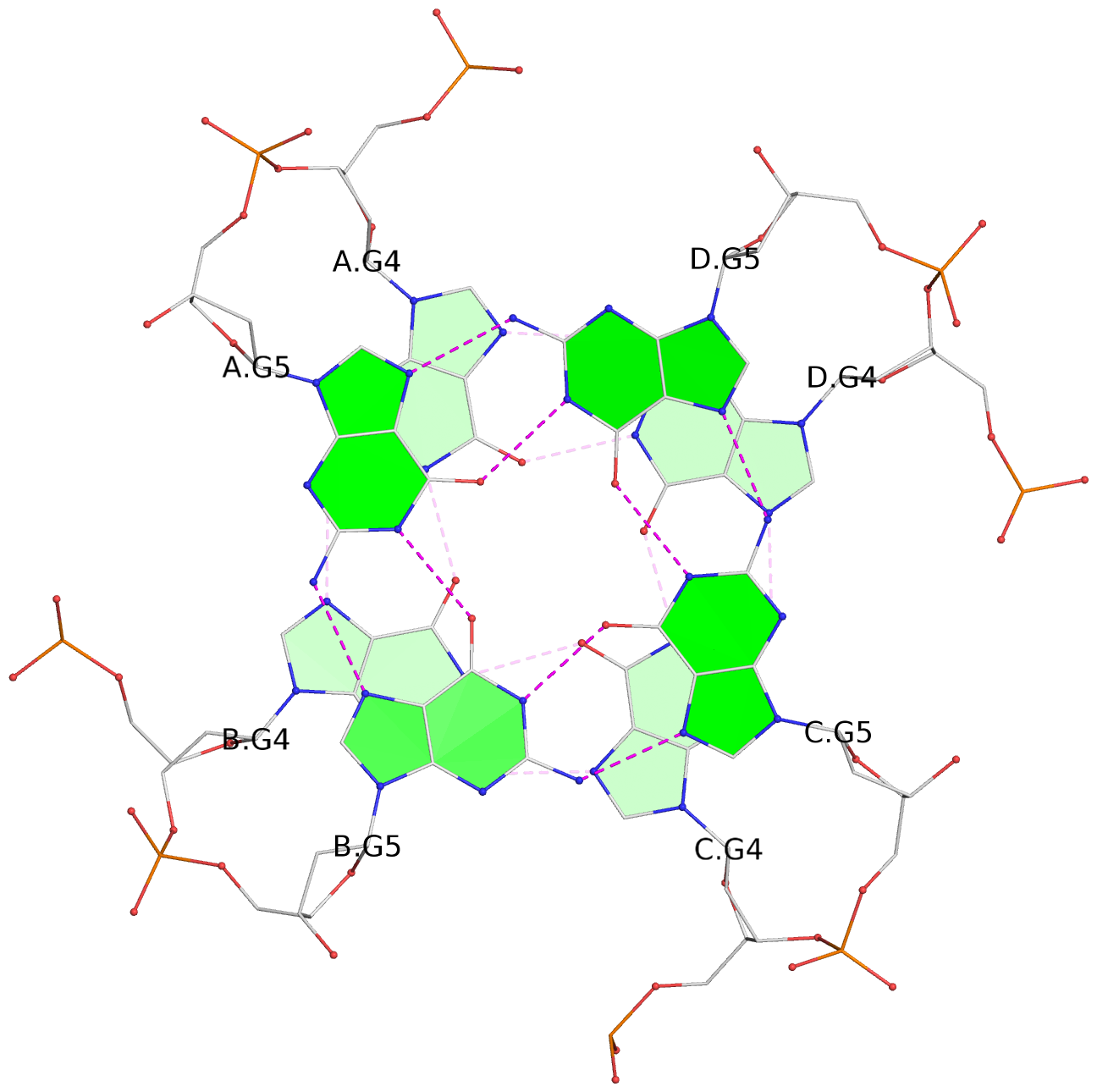
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, parallel(4+0), UUUU
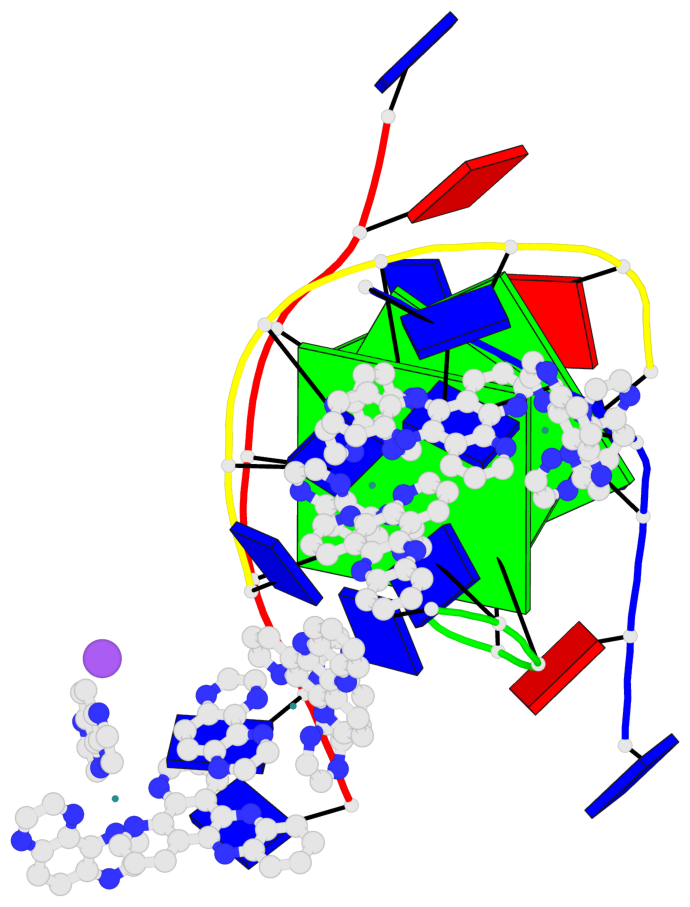
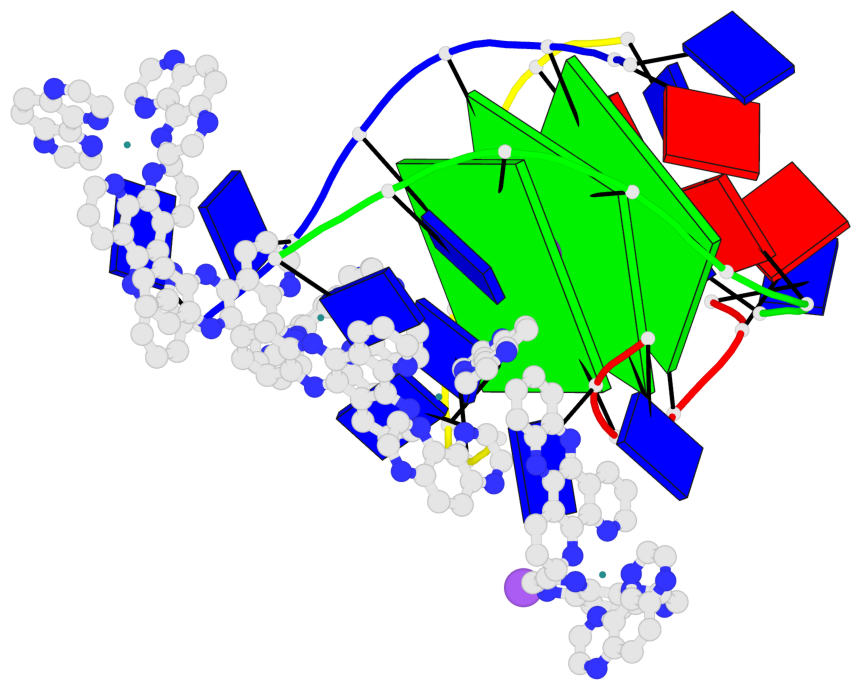
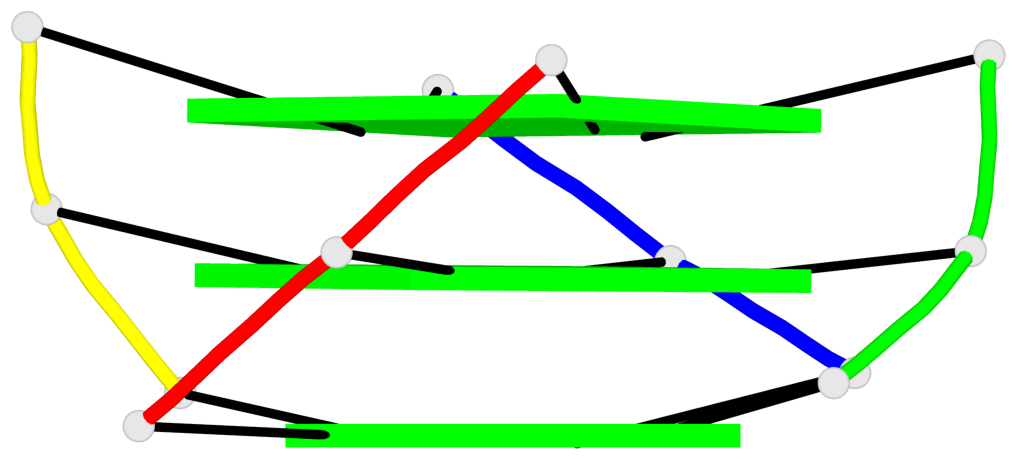
Base-block schematics in six views
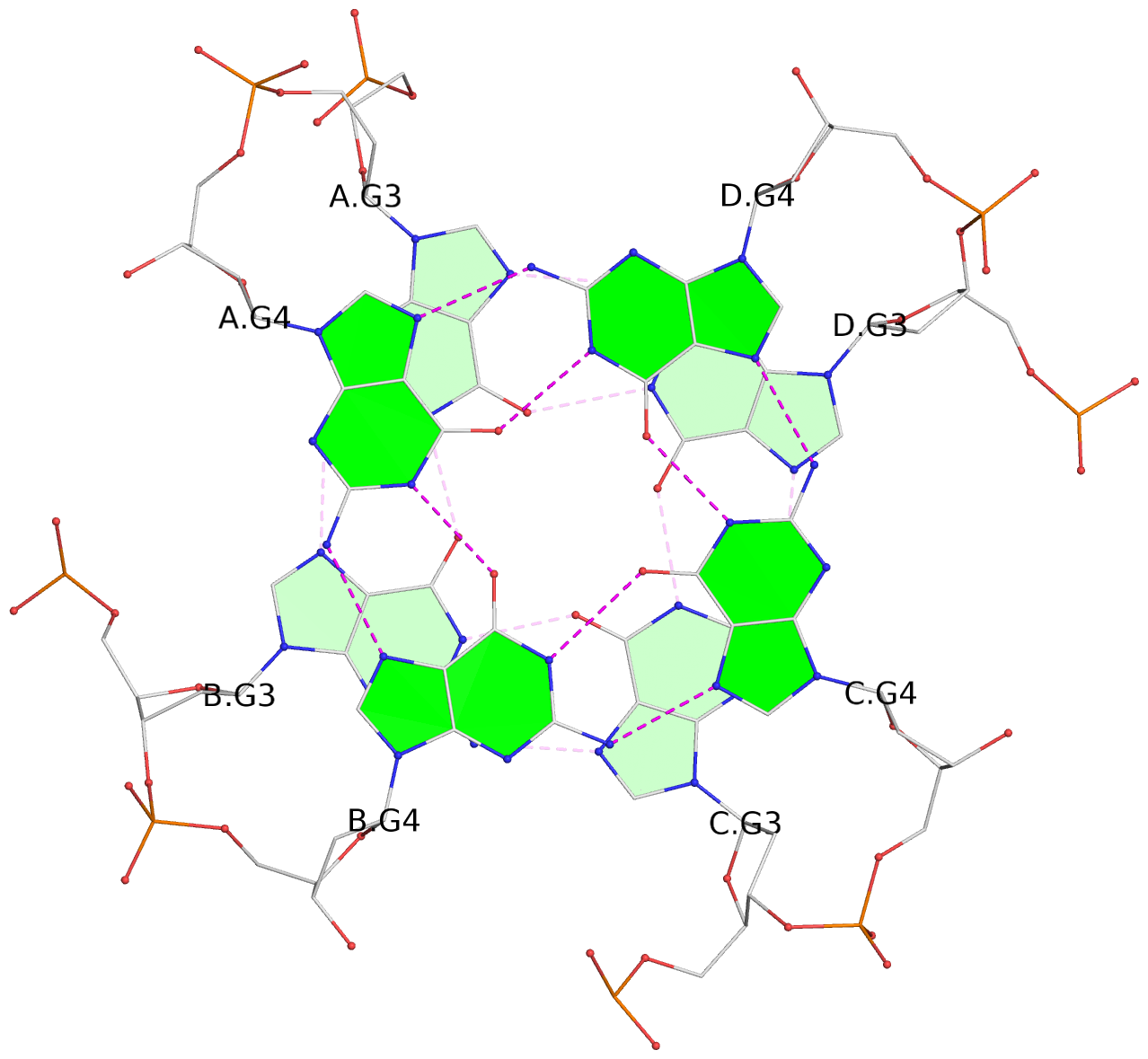
List of 3 G-tetrads
1 glyco-bond=---- sugar=--3- groove=---- planarity=0.175 type=other nts=4 GGGG A.DG3,B.DG3,C.DG3,D.DG3 2 glyco-bond=---- sugar=---- groove=---- planarity=0.192 type=other nts=4 GGGG A.DG4,B.DG4,C.DG4,D.DG4 3 glyco-bond=---- sugar=---- groove=---- planarity=0.302 type=bowl nts=4 GGGG A.DG5,B.DG5,C.DG5,D.DG5
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, inter-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.