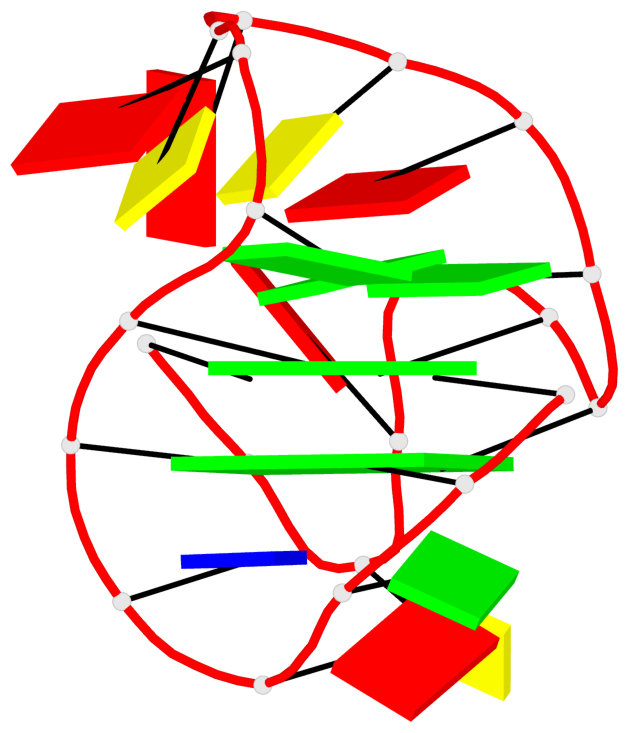
Detailed DSSR results for the G-quadruplex: PDB entry 6rs3
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 6rs3
- Class
- DNA
- Method
- NMR
- Summary
- 2'-f-riboguanosine modified g-quadruplex with v-loop
- Reference
- Haase L, Dickerhoff J, Weisz K (2020): "Sugar Puckering Drives G-Quadruplex Refolding: Implications for V-Shaped Loops." Chemistry, 26, 524-533. doi: 10.1002/chem.201904044.
- Abstract
- A DNA G-quadruplex adopting a (3+1) hybrid structure was modified in two adjacent syn positions of the antiparallel strand with anti-favoring 2'-deoxy-2'-fluoro-riboguanosine (F rG) analogues. The two substitutions promoted a structural rearrangement to a topology with the 5'-terminal G residue located in the central tetrad and the two modified residues linked by a V-shaped zero-nucleotide loop. Strikingly, whereas a sugar pucker in the preferred north domain is found for both modified nucleotides, the F rG analogue preceding the V-loop is forced to adopt the unfavored syn conformation in the new quadruplex fold. Apparently, a preferred C3'-endo sugar pucker within the V-loop architecture outweighs the propensity of the F rG analogue to adopt an anti glycosidic conformation. Refolding into a V-loop topology is likewise observed for a sequence modified at corresponding positions with two riboguanosine substitutions. In contrast, 2'-F-arabinoguanosine analogues with their favored south-east sugar conformation do not support formation of the V-loop topology. Examination of known G-quadruplexes with a V-shaped loop highlights the critical role of the sugar conformation for this distinct structural motif.
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 2(-LwX+P), UD3(1+3), UDDD
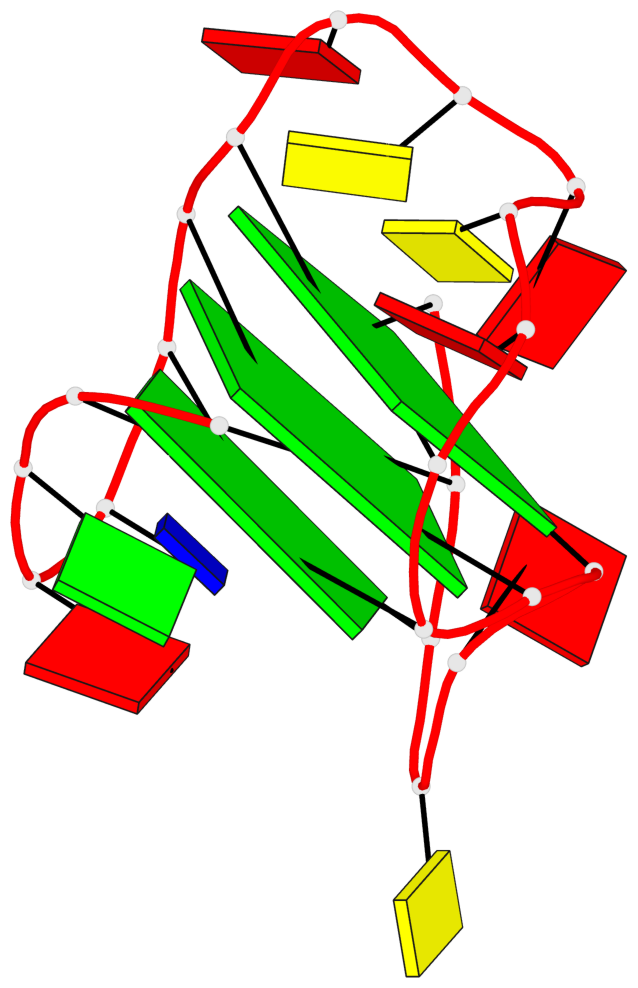
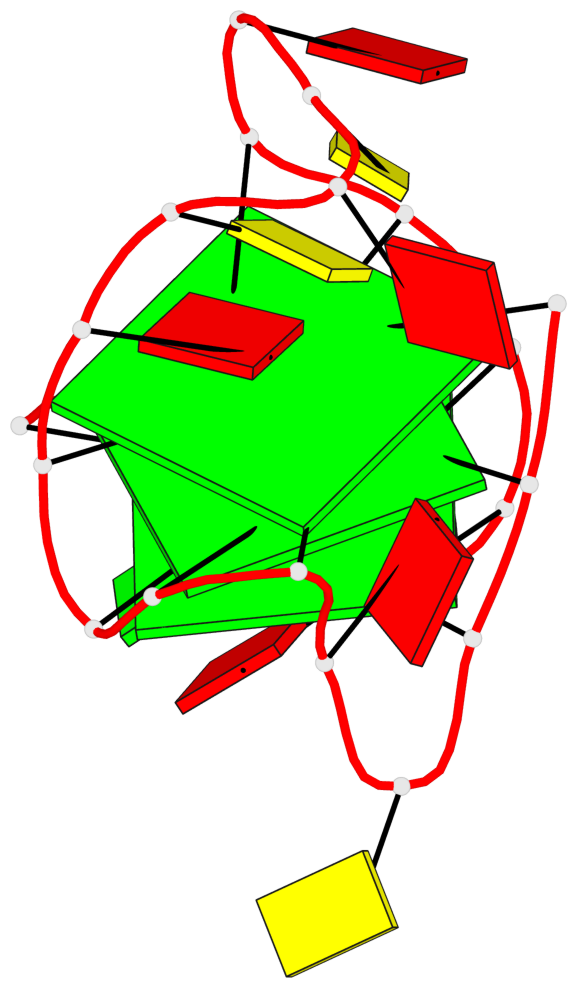
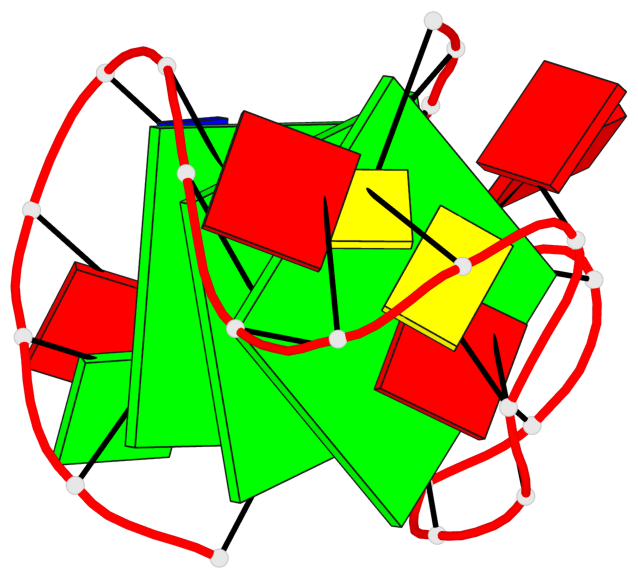
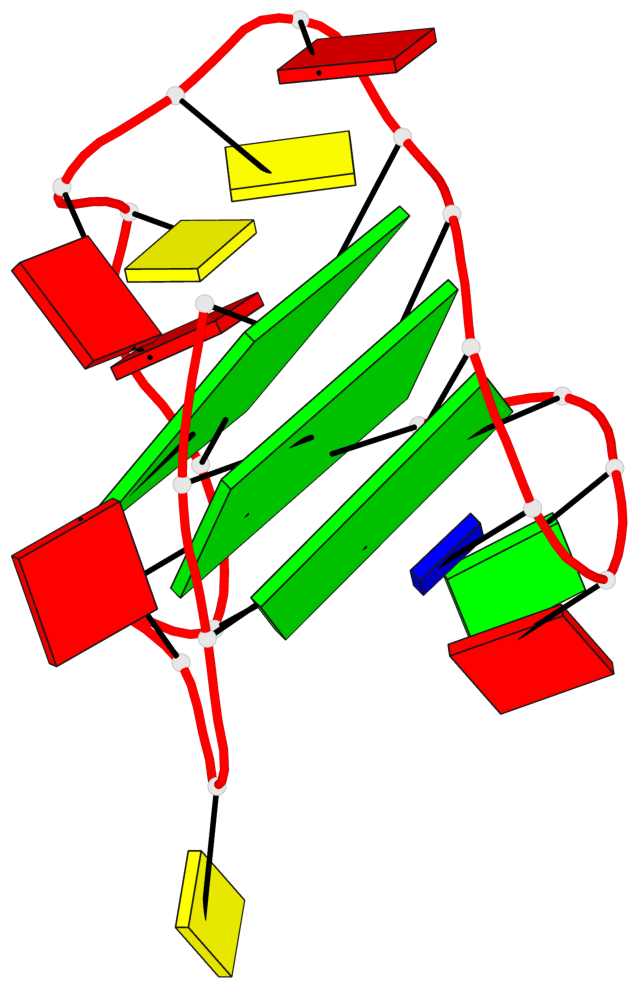
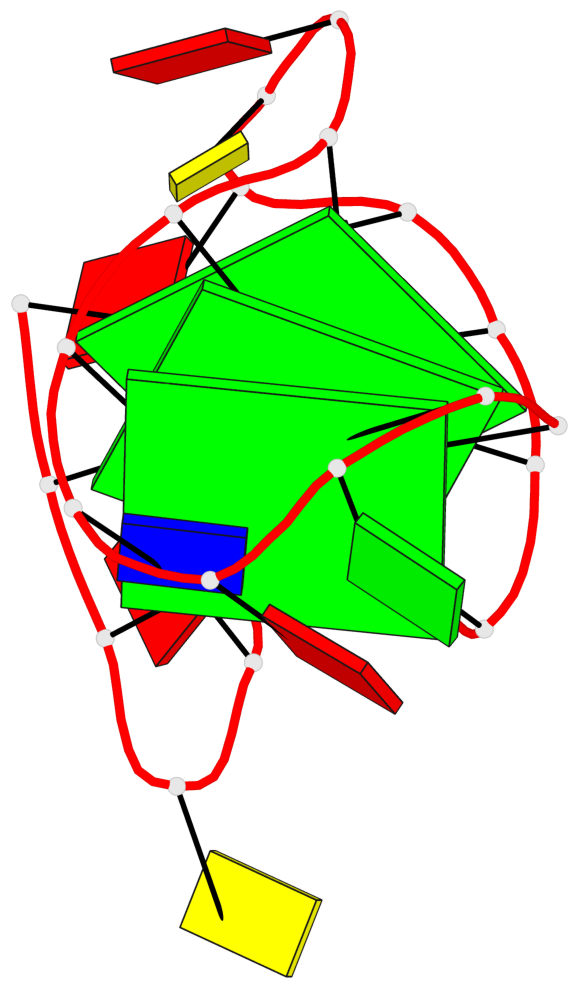
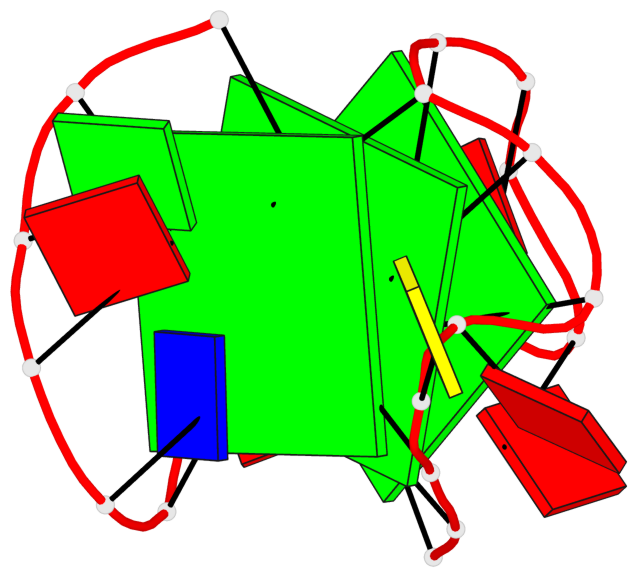
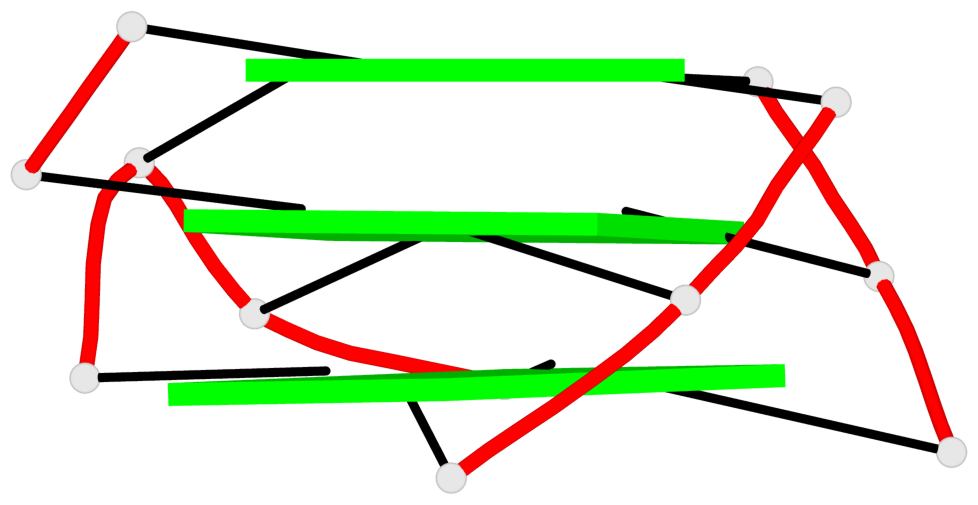
Base-block schematics in six views
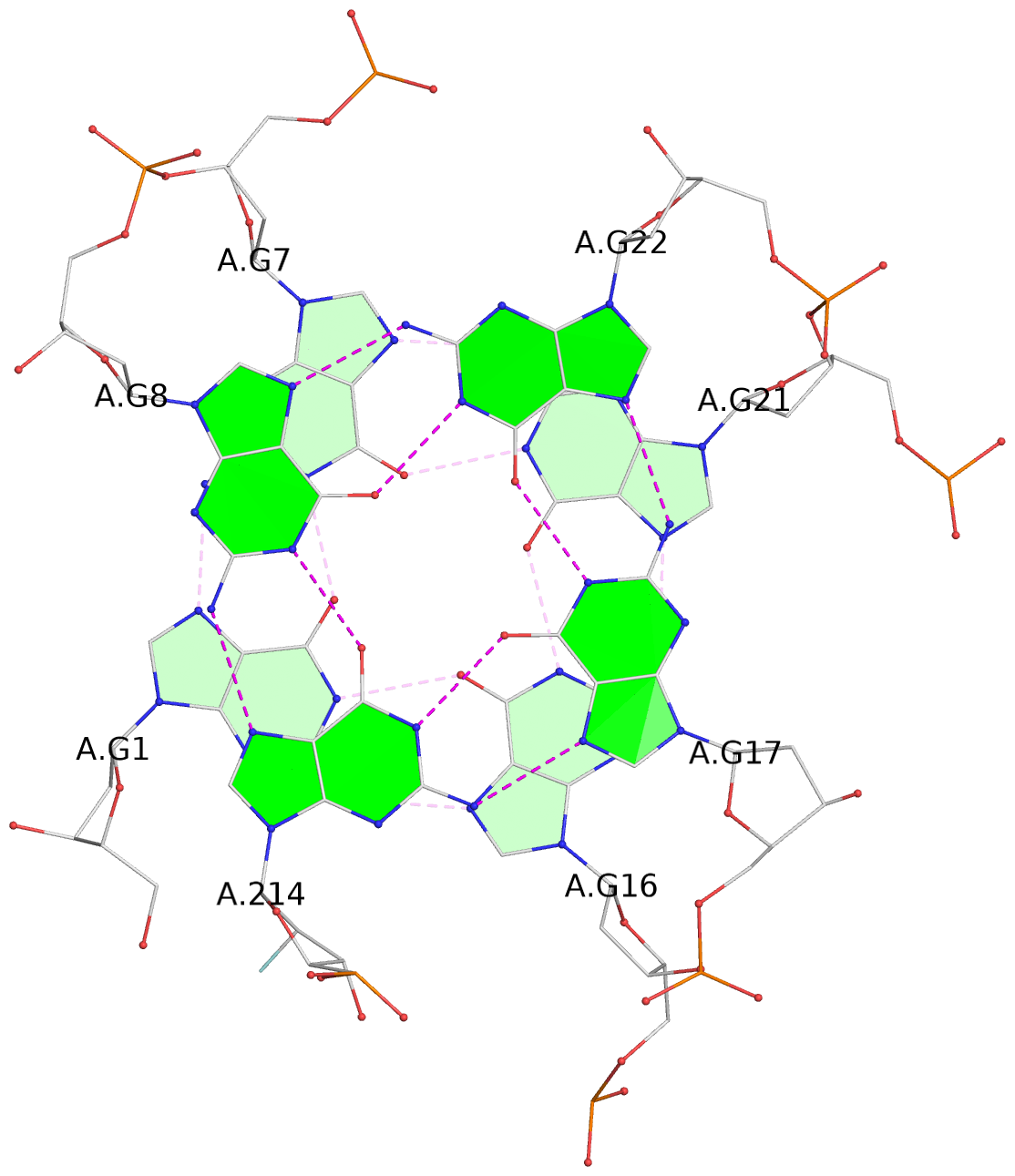
List of 3 G-tetrads
1 glyco-bond=s--- sugar=---- groove=w--n planarity=0.407 type=bowl-2 nts=4 GGGG A.DG1,A.DG7,A.DG21,A.DG16 2 glyco-bond=-ss- sugar=---. groove=w-n- planarity=0.364 type=saddle nts=4 GGGg A.DG2,A.DG6,A.DG20,A.GF2/15 3 glyco-bond=-s-- sugar=-3-- groove=wn-- planarity=0.354 type=other nts=4 GgGG A.DG8,A.GF2/14,A.DG17,A.DG22
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 2 G-tetrad layers, 3 loops, INTRA-molecular, UDDD, hybrid-(mixed), 2(-LwX+P), UD3(1+3)
List of 2 non-stem G4-loops (including the two closing Gs)
1 type=lateral helix=#1 nts=7 GACACAg A.DG8,A.DA9,A.DC10,A.DA11,A.DC12,A.DA13,A.GF2/14 2 type=V-shaped helix=#1 nts=4 ggGG A.GF2/14,A.GF2/15,A.DG16,A.DG17