Detailed DSSR results for the G-quadruplex: PDB entry 6s15
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 6s15
- Class
- DNA
- Method
- X-ray (1.7 Å)
- Summary
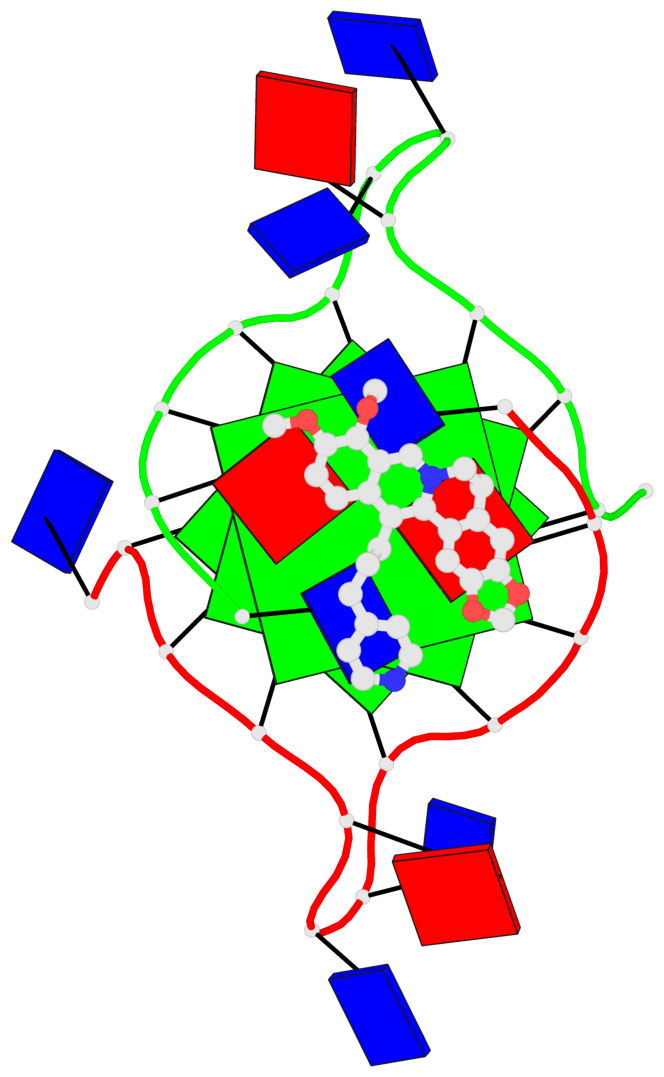
- Pyridine derivative of the natural alkaloid berberine as human telomeric g-quadruplex binder
- Reference
- Papi F, Bazzicalupi C, Ferraroni M, Ciolli G, Lombardi P, Khan AY, Kumar GS, Gratteri P (2020): "Pyridine Derivative of the Natural Alkaloid Berberine as Human Telomeric G4-DNA Binder: A Solution and Solid-State Study." Acs Med.Chem.Lett., 11, 645-650. doi: 10.1021/acsmedchemlett.9b00516.
- Abstract
- Telomerase is an enzyme deputed to the maintenance of eukaryotic chromosomes; however, its overexpression is a recognized hallmark of many cancer forms. A viable route for the inhibition of telomerase in malignant cells is the stabilization of G-quadruplex structures (G4) at the 3' overhang of telomeres. Berberine has shown in this regard valuable G4 binding properties together with a significant anticancer activity and telomerase inhibition effects. Here, we focused on a berberine derivative featuring a pyridine containing side group at the 13th position. Such modification actually improves the binding toward telomeric G-quadruplexes and establishes a degree of selectivity in the interaction with different sequences. Moreover, the X-ray crystal structure obtained for the complex formed by the ligand and a bimolecular human telomeric quadruplex affords a better understanding of the 13-berberine derivatives behavior with telomeric G4 and allows to draw useful insights for the future design of derivatives with remarkable anticancer properties.
- G4 notes
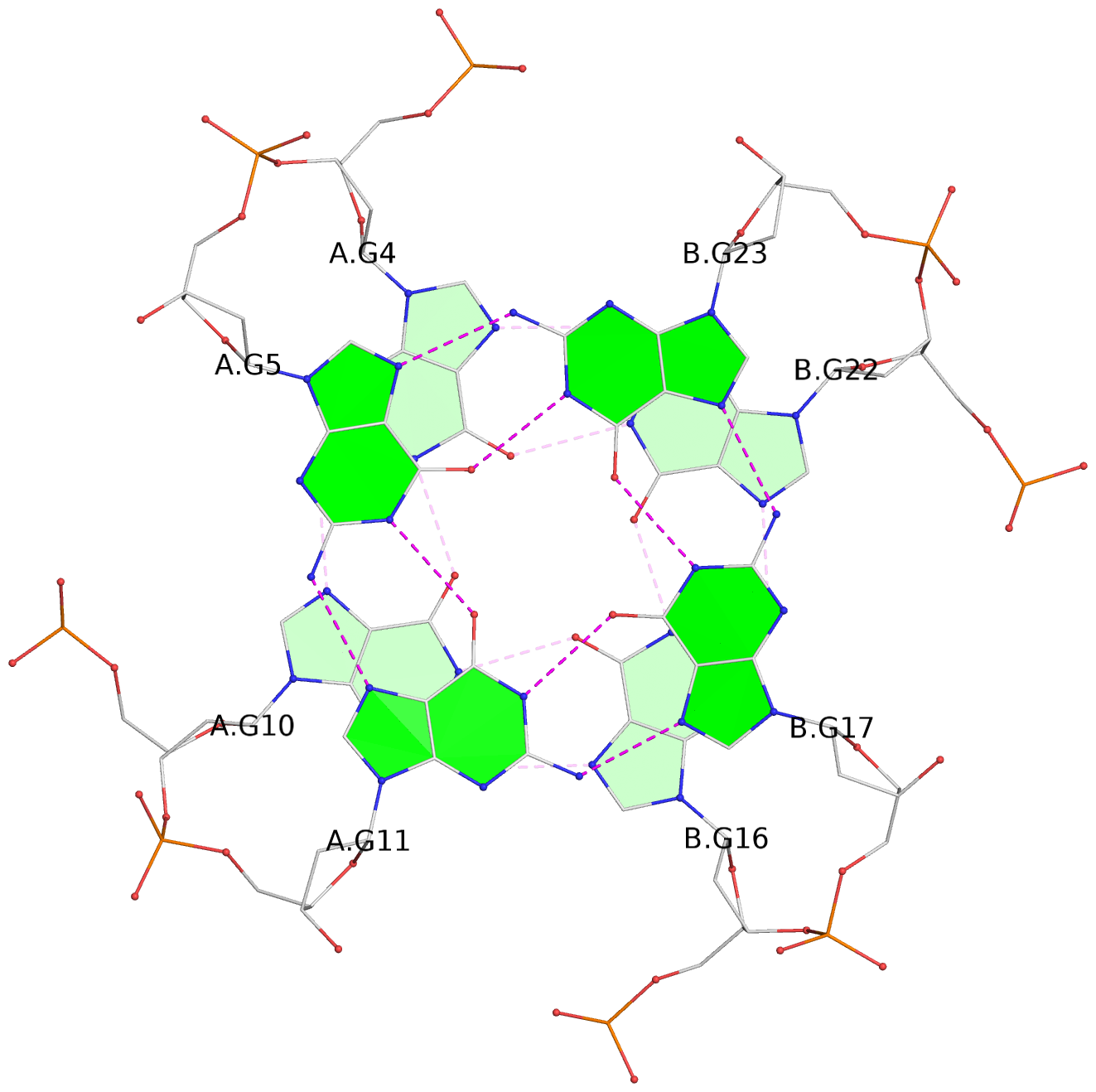
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, parallel(4+0), UUUU
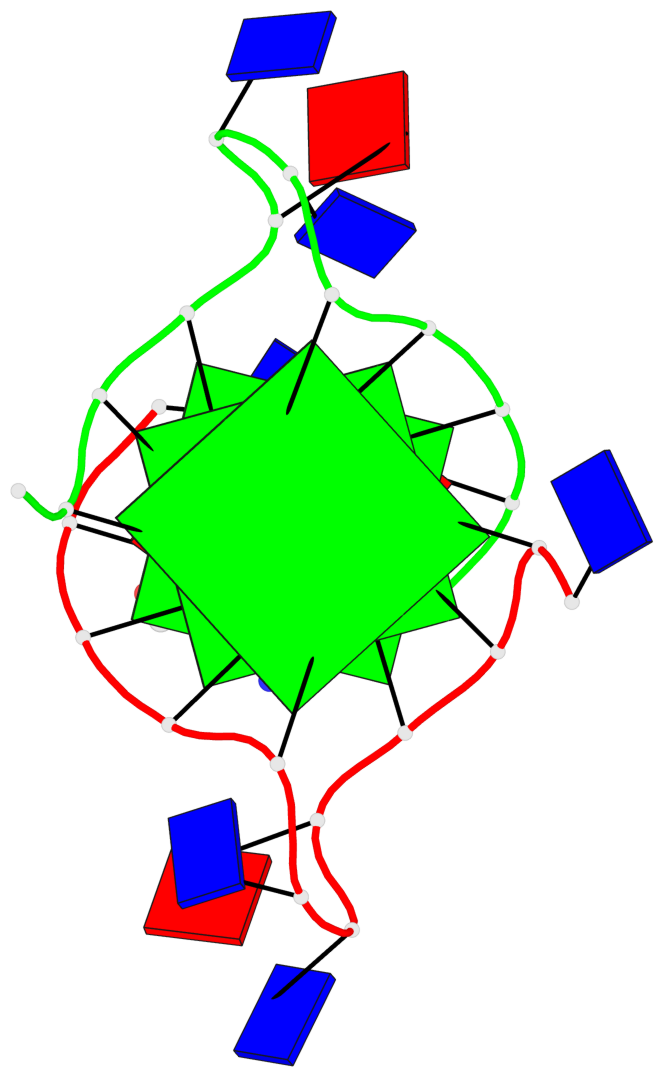
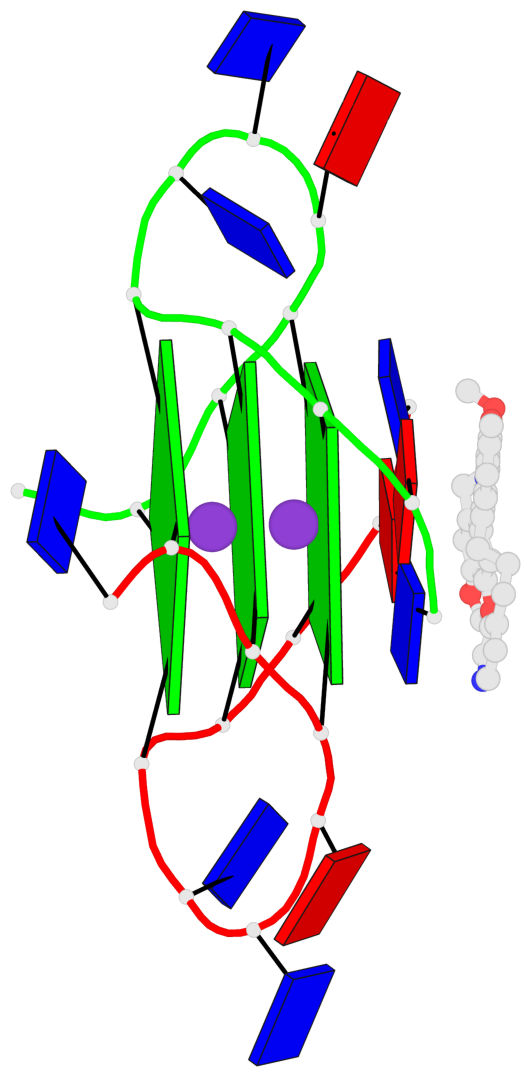
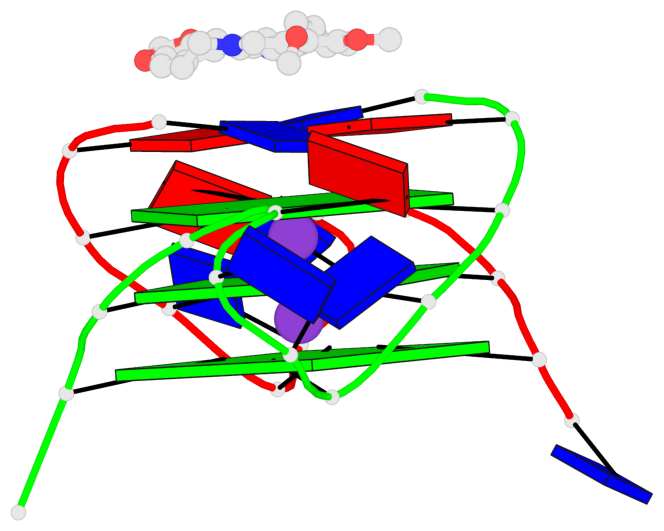
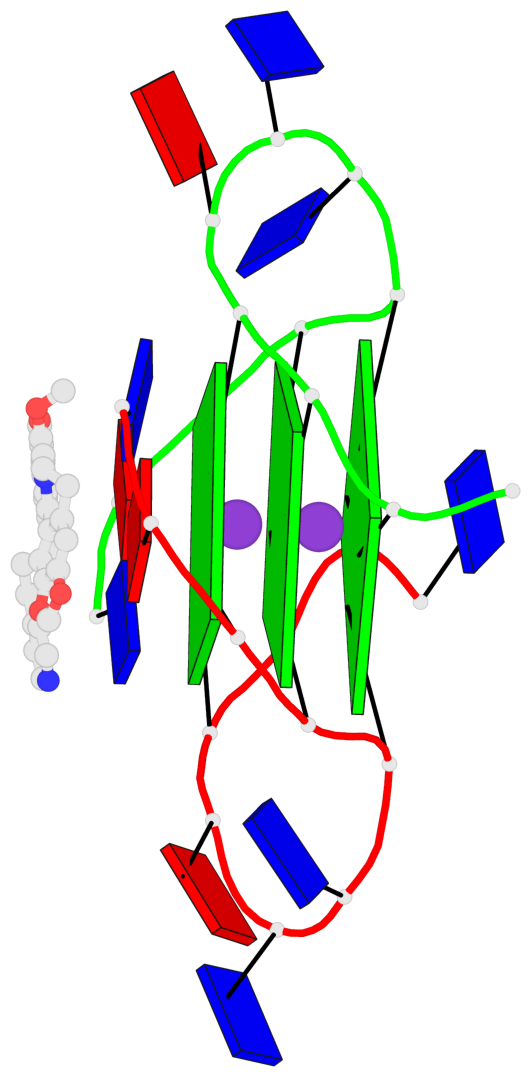
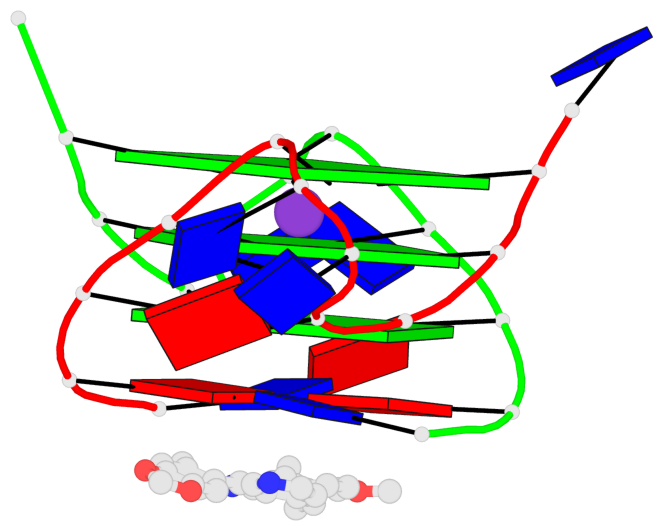
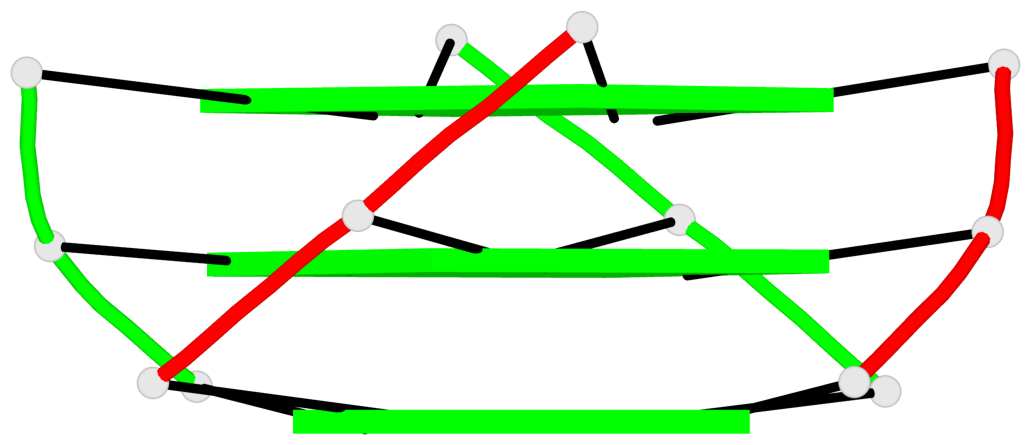
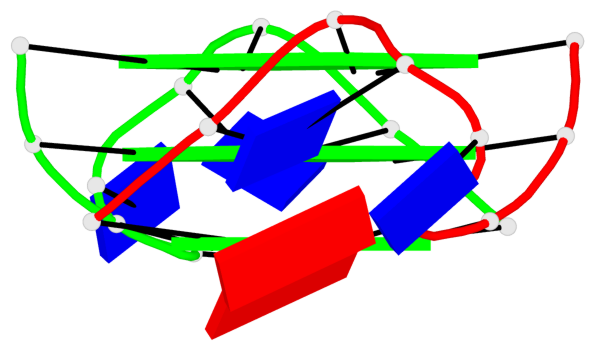
Base-block schematics in six views
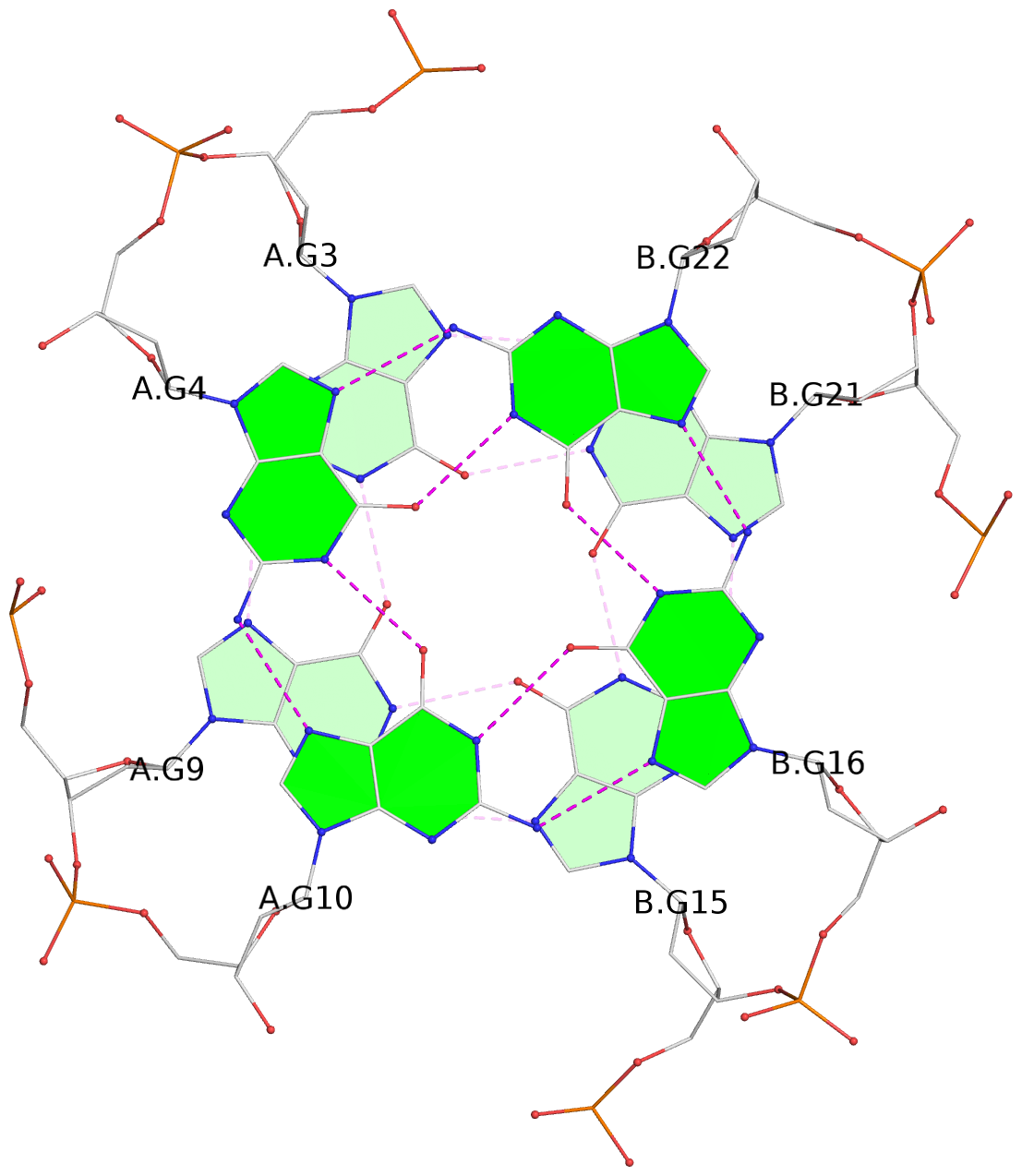
List of 3 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.109 type=planar nts=4 GGGG A.DG3,A.DG9,B.DG15,B.DG21 2 glyco-bond=---- sugar=---- groove=---- planarity=0.144 type=planar nts=4 GGGG A.DG4,A.DG10,B.DG16,B.DG22 3 glyco-bond=---- sugar=---- groove=---- planarity=0.306 type=bowl nts=4 GGGG A.DG5,A.DG11,B.DG17,B.DG23
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, inter-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.