Detailed DSSR results for the G-quadruplex: PDB entry 6suu
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 6suu
- Class
- DNA
- Method
- NMR
- Summary
- NMR structure of kras32r g9t conformer g-quadruplex within kras promoter region
- Reference
- Marquevielle J, Robert C, Lagrabette O, Wahid M, Bourdoncle A, Xodo LE, Mergny JL, Salgado GF (2020): "Structure of two G-quadruplexes in equilibrium in the KRAS promoter." Nucleic Acids Res., 48, 9336-9345. doi: 10.1093/nar/gkaa387.
- Abstract
- KRAS is one of the most mutated oncogenes and still considered an undruggable target. An alternative strategy would consist in targeting its gene rather than the protein, specifically the formation of G-quadruplexes (G4) in its promoter. G4 are secondary structures implicated in biological processes, which can be formed among G-rich DNA (or RNA) sequences. Here we have studied the major conformations of the commonly known KRAS 32R, or simply 32R, a 32 residue sequence within the KRAS Nuclease Hypersensitive Element (NHE) region. We have determined the structure of the two major stable conformers that 32R can adopt and which display slow equilibrium (>ms) with each other. By using different biophysical methods, we found that the nucleotides G9, G25, G28 and G32 are particularly implicated in the exchange between these two conformations. We also showed that a triad at the 3' end further stabilizes one of the G4 conformations, while the second conformer remains more flexible and less stable.
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 2(-P-P-P), parallel(4+0), UUUU
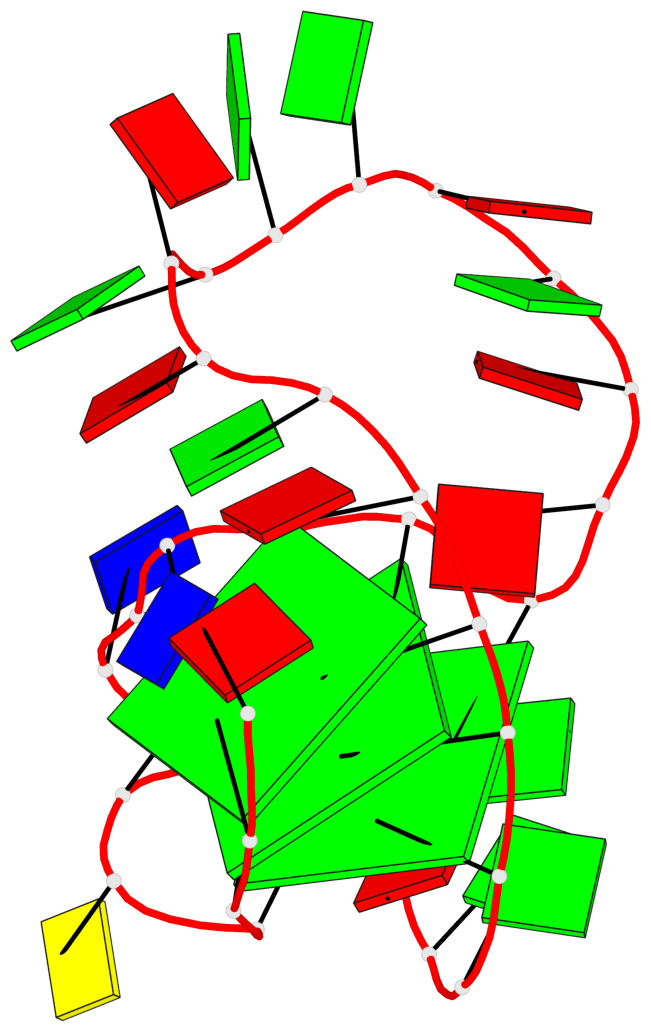
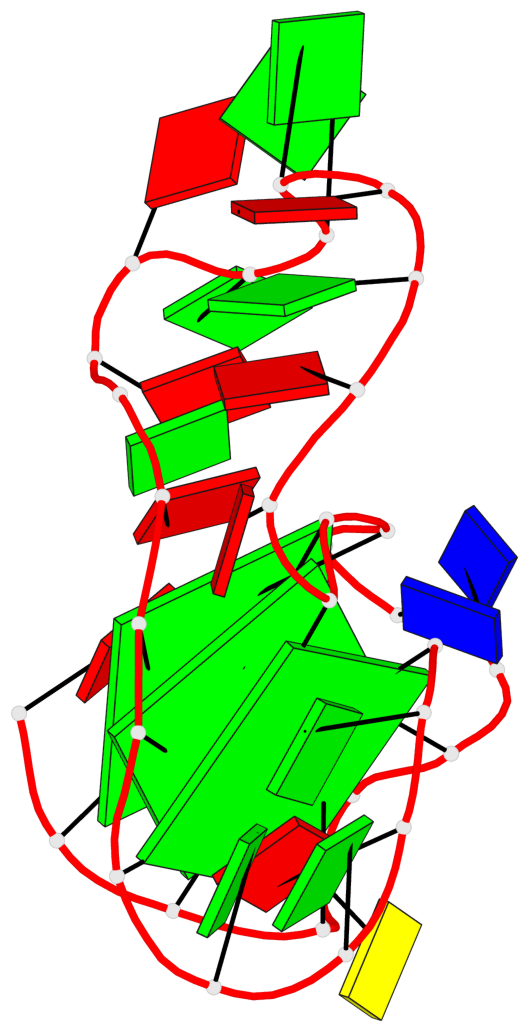
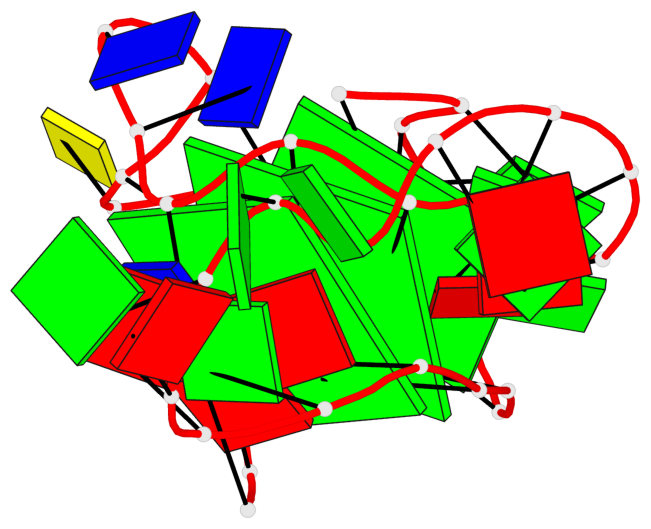
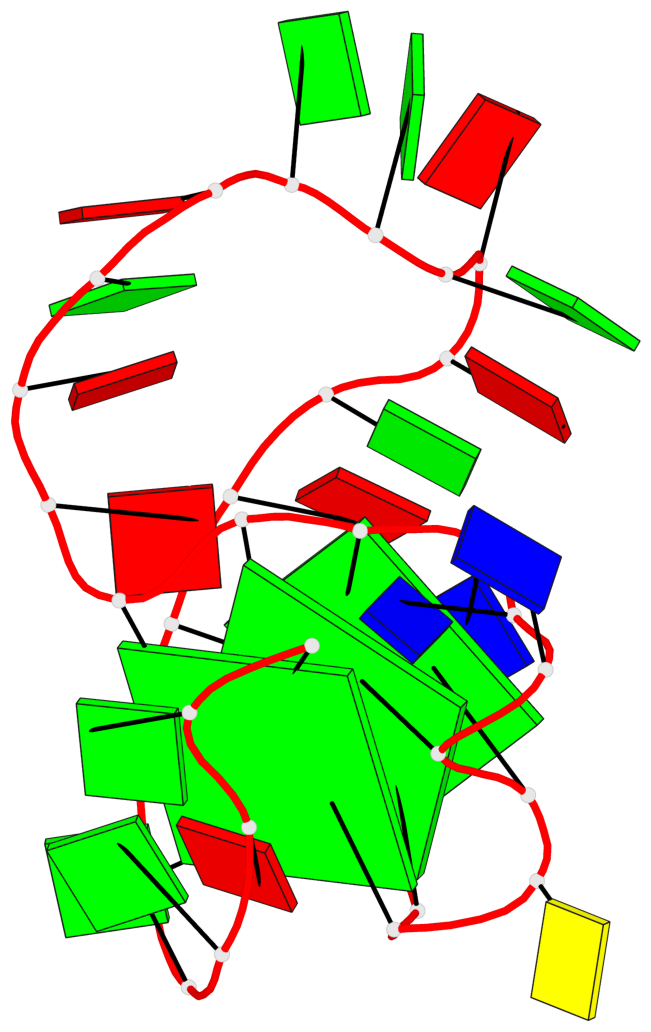
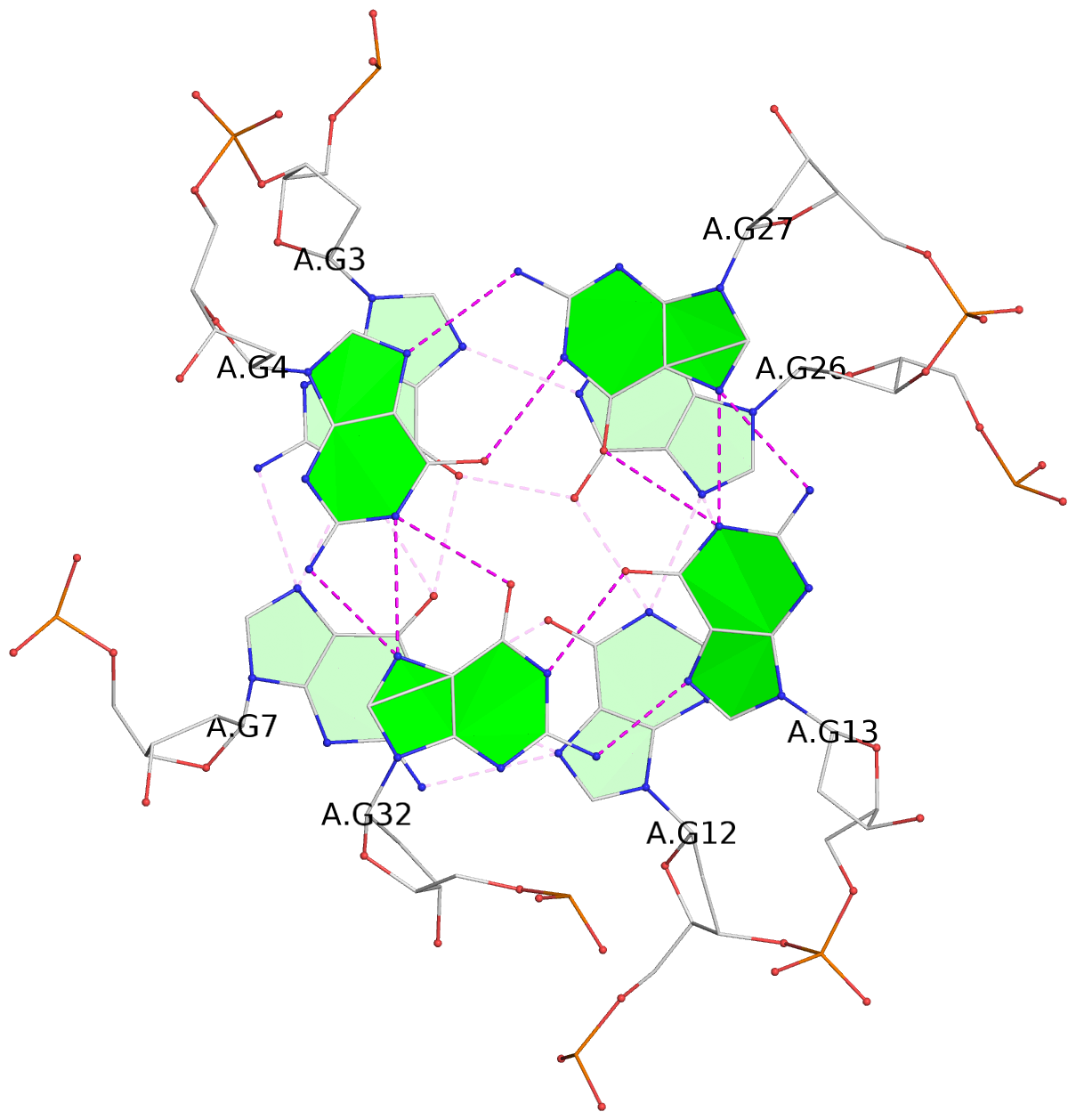
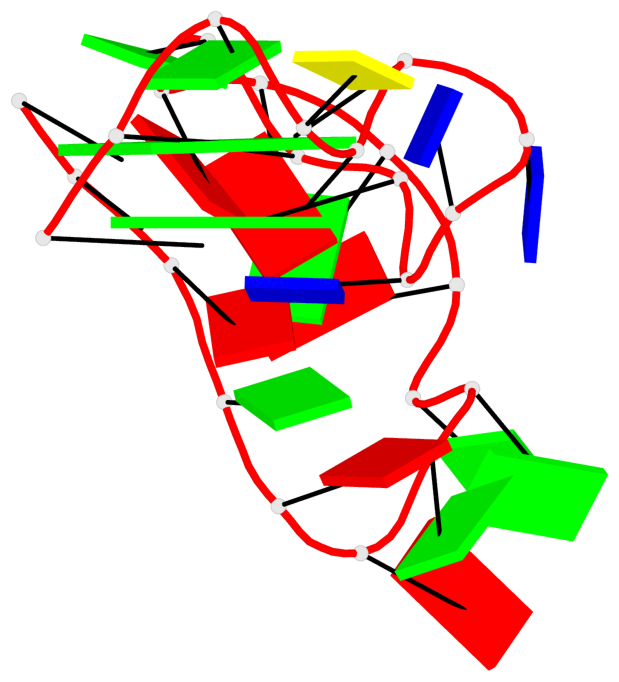
Base-block schematics in six views
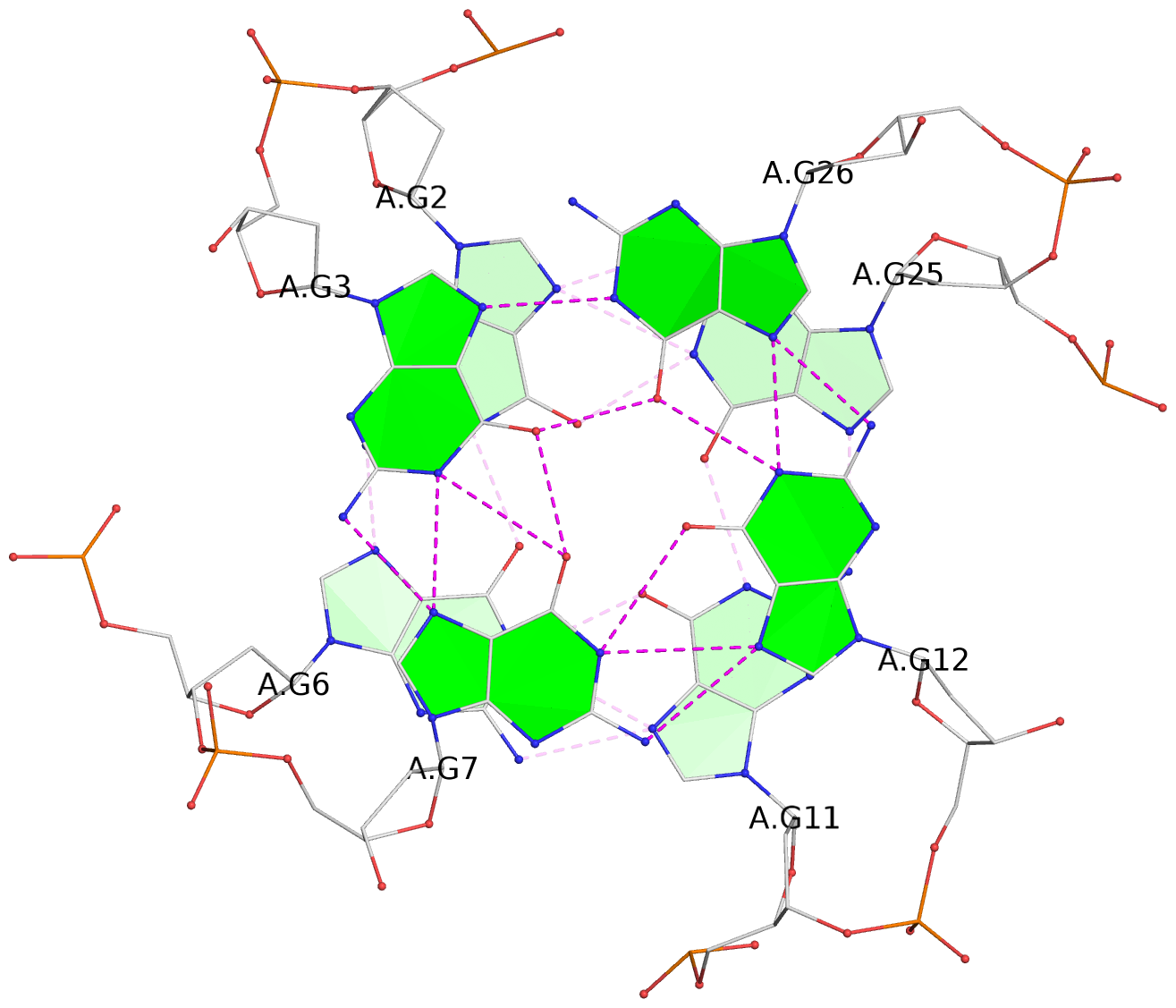
List of 3 G-tetrads
1 glyco-bond=---- sugar=--.. groove=---- planarity=0.677 type=bowl nts=4 GGGG A.DG2,A.DG6,A.DG11,A.DG25 2 glyco-bond=---- sugar=..-3 groove=---- planarity=0.602 type=other nts=4 GGGG A.DG3,A.DG7,A.DG12,A.DG26 3 glyco-bond=-s-- sugar=-..- groove=wn-- planarity=0.507 type=saddle nts=4 GGGG A.DG4,A.DG32,A.DG13,A.DG27
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(-P-P-P), parallel(4+0)
List of 1 non-stem G4-loop (including the two closing Gs)
1 type=diagonal helix=#1 nts=6 GGGAGG A.DG27,A.DG28,A.DG29,A.DA30,A.DG31,A.DG32