Detailed DSSR results for the G-quadruplex: PDB entry 6syk
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 6syk
- Class
- DNA
- Method
- NMR
- Summary
- Guanine-rich oligonucleotide with 5'- and 3'-gc ends form g-quadruplex with a(gggg)a hexad, gcgc- and g-quartets and two symmetric gg and aa base pair
- Reference
- Pavc D, Wang B, Spindler L, Drevensek-Olenik I, Plavec J, Sket P (2020): "GC ends control topology of DNA G-quadruplexes and their cation-dependent assembly." Nucleic Acids Res., 48, 2749-2761. doi: 10.1093/nar/gkaa058.
- Abstract
- GCn and GCnCG, where n = (G2AG4AG2), fold into well-defined, dimeric G-quadruplexes with unprecedented folding topologies in the presence of Na+ ions as revealed by nuclear magnetic resonance spectroscopy. Both G-quadruplexes exhibit unique combination of structural elements among which are two G-quartets, A(GGGG)A hexad and GCGC-quartet. Detailed structural characterization uncovered the crucial role of 5'-GC ends in formation of GCn and GCnCG G-quadruplexes. Folding in the presence of 15NH4+ and K+ ions leads to 3'-3' stacking of terminal G-quartets of GCn G-quadruplexes, while 3'-GC overhangs in GCnCG prevent dimerization. Results of the present study expand repertoire of possible G-quadruplex structures. This knowledge will be useful in DNA sequence design for nanotechnological applications that may require specific folding topology and multimerization properties.
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, parallel(4+0), UUUU
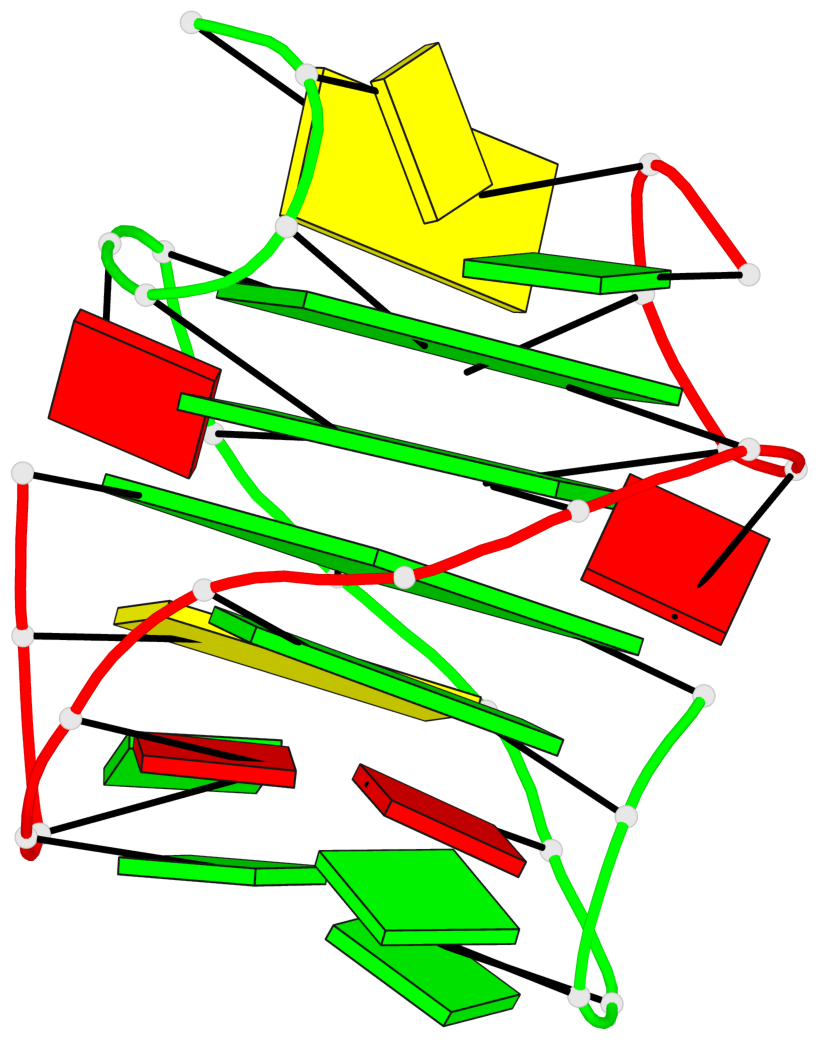
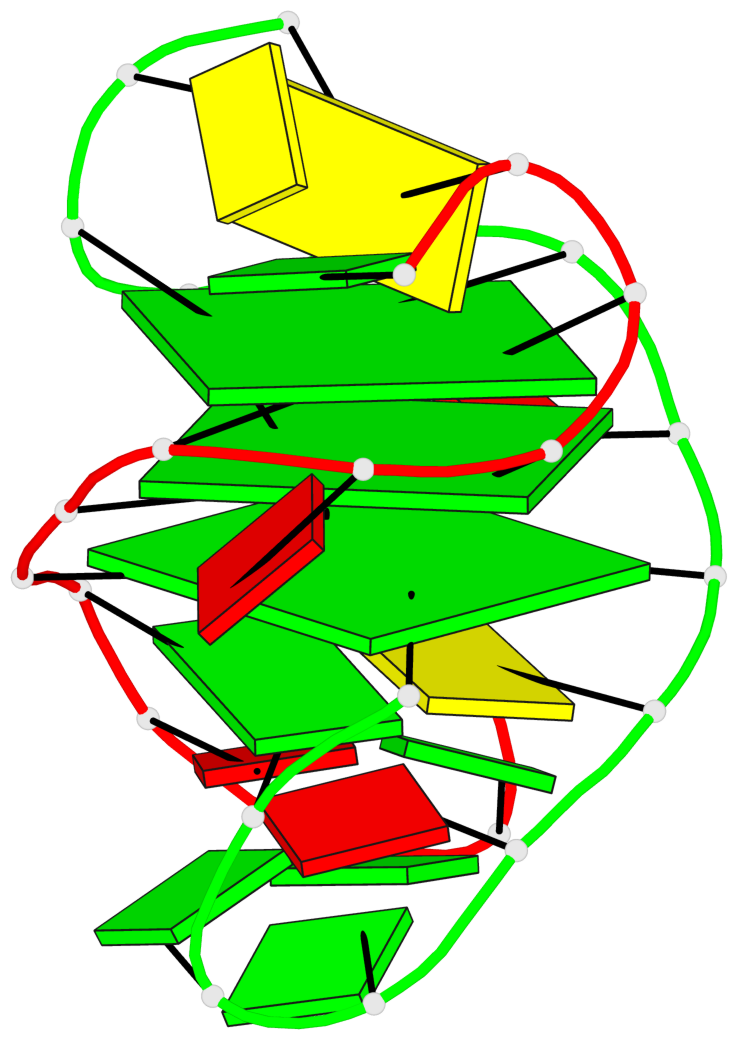
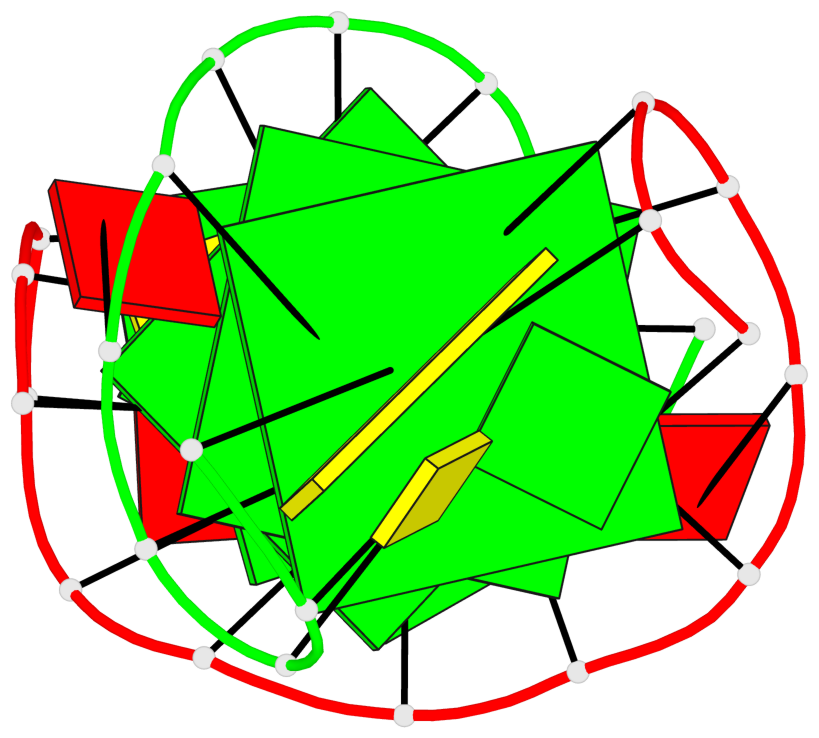
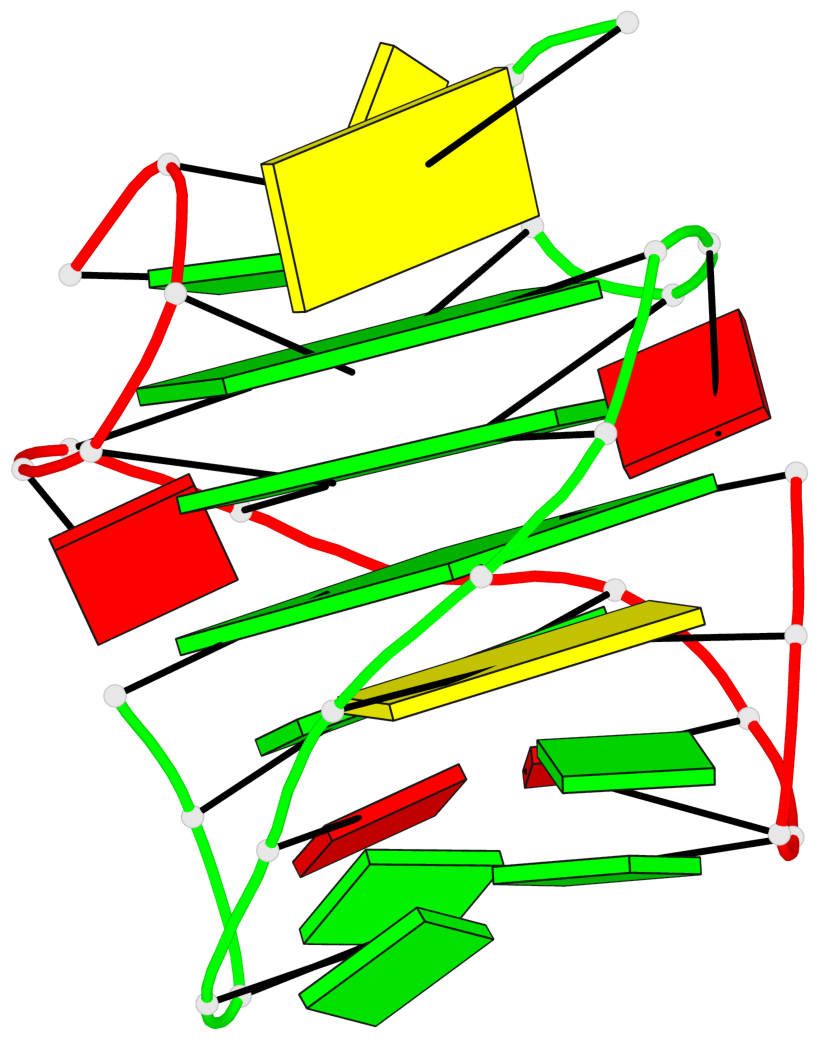
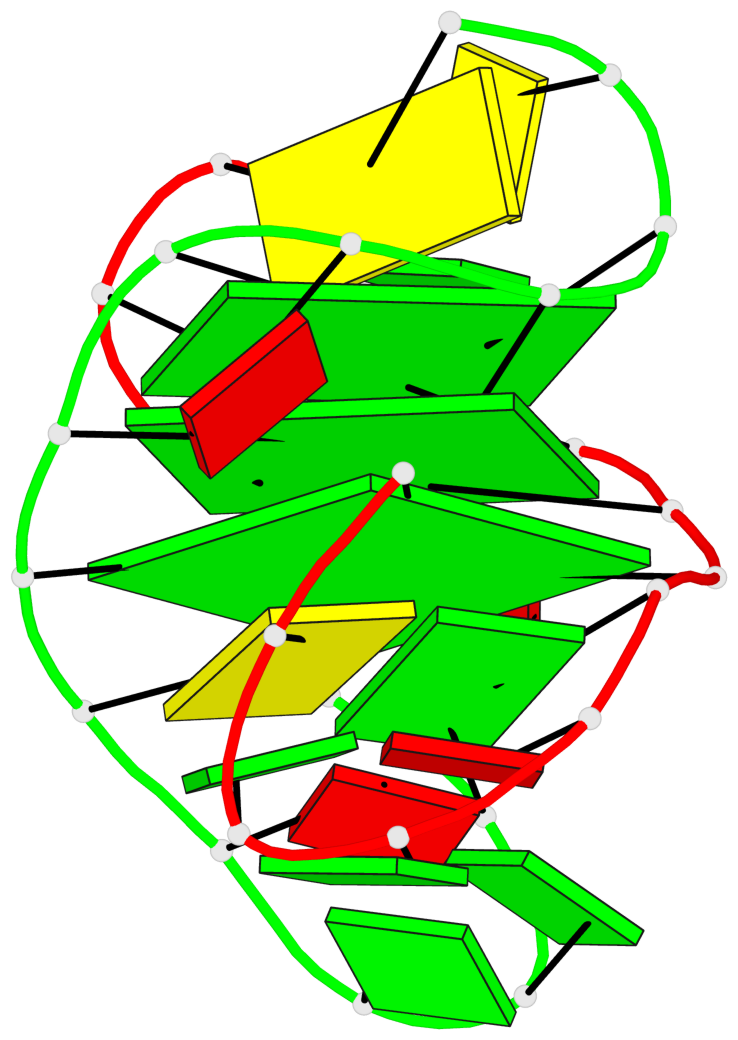
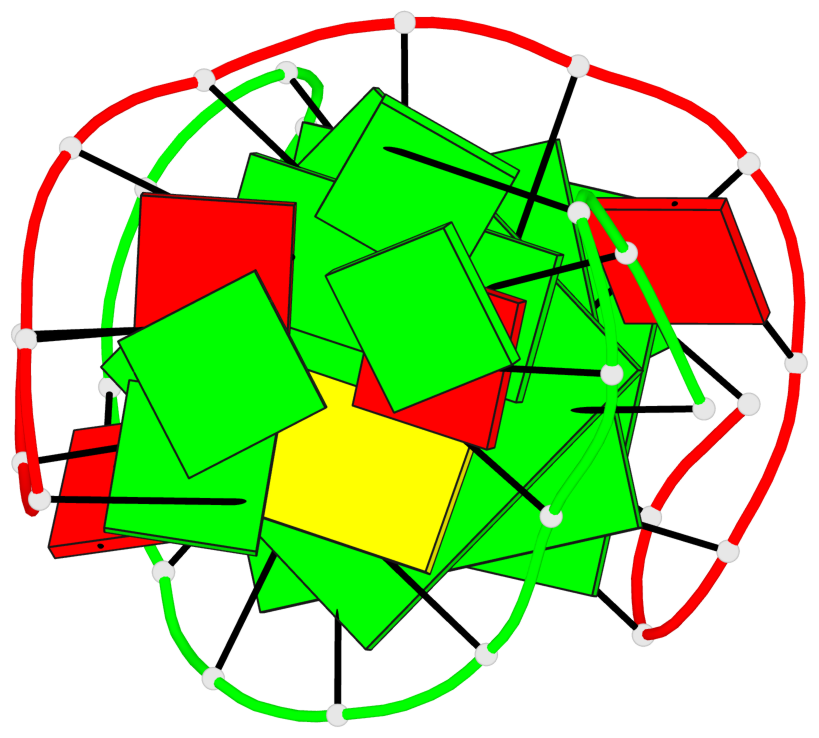
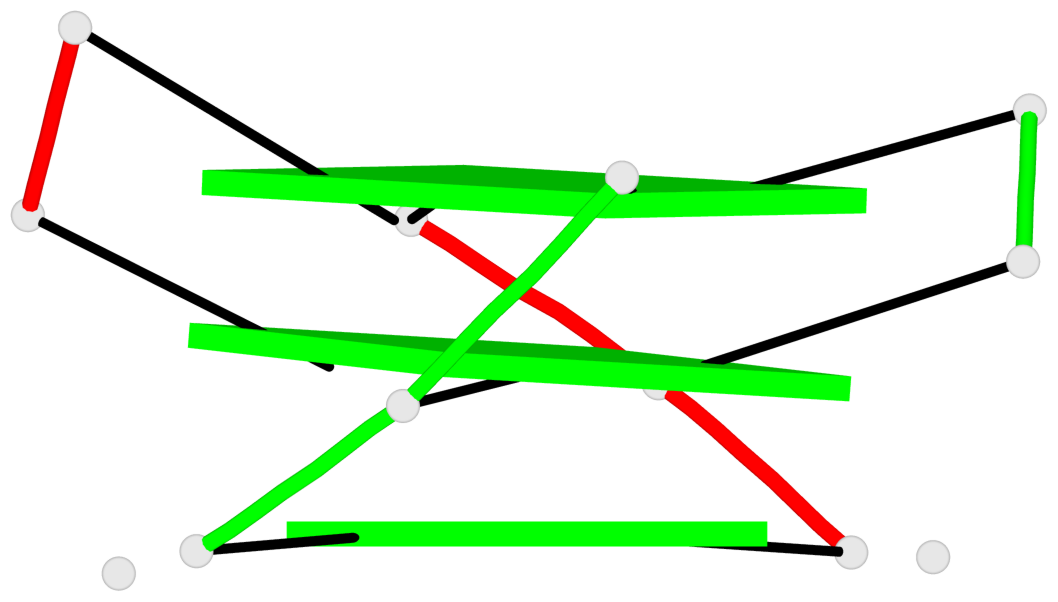
Base-block schematics in six views
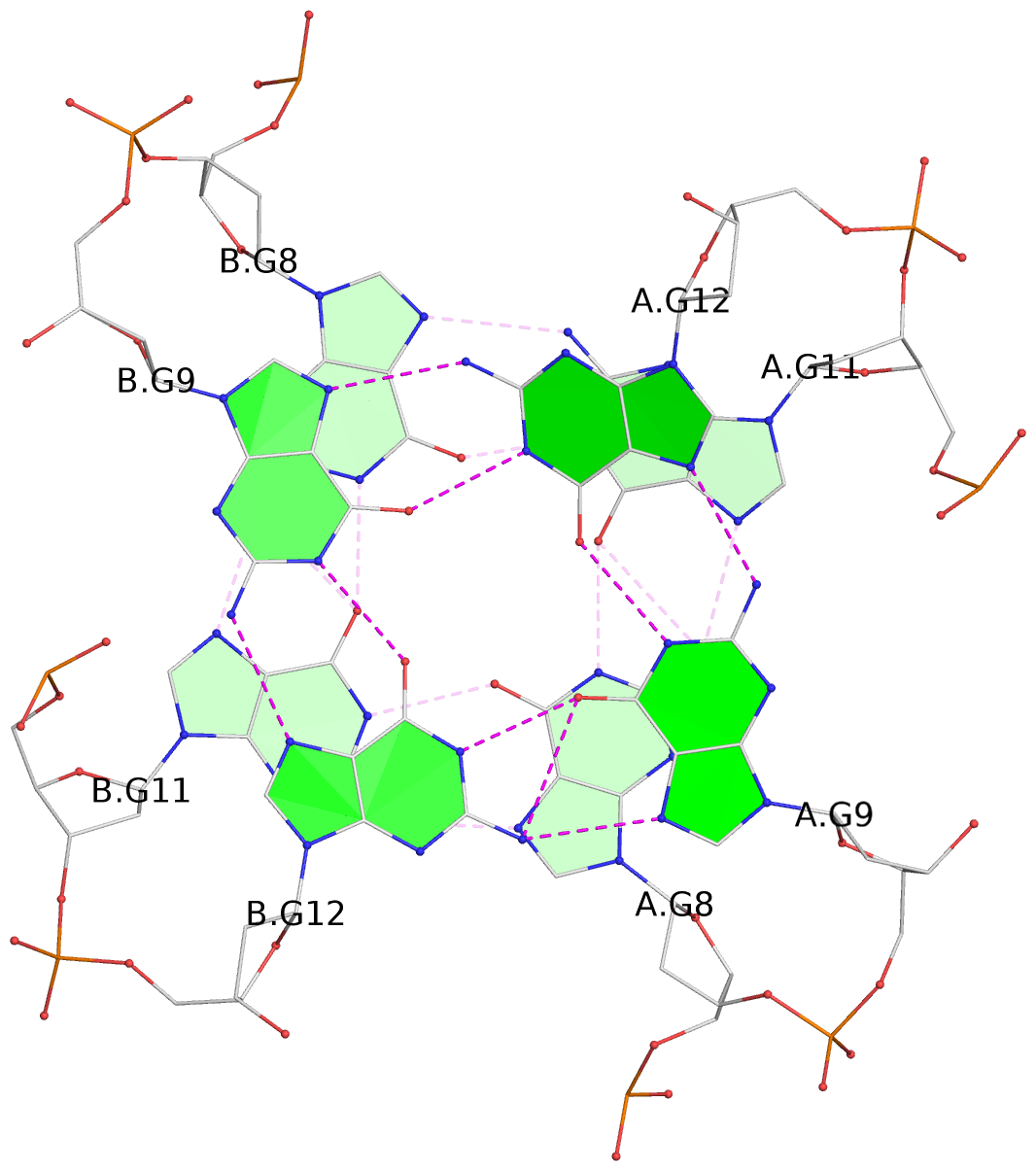
List of 3 G-tetrads
1 glyco-bond=s-s- sugar=---- groove=wnwn planarity=0.291 type=other nts=4 GGGG A.DG1,B.DG7,B.DG1,A.DG7 2 glyco-bond=---- sugar=---- groove=---- planarity=0.509 type=bowl-2 nts=4 GGGG A.DG8,A.DG11,B.DG8,B.DG11 3 glyco-bond=---- sugar=---- groove=---- planarity=0.625 type=bowl nts=4 GGGG A.DG9,A.DG12,B.DG9,B.DG12
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, inter-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 2 G-tetrad layers, 2 loops, inter-molecular, UUUU, parallel, parallel(4+0)
List of 2 non-stem G4-loops (including the two closing Gs)
1 type=lateral helix=#1 nts=7 GCGGAGG A.DG1,A.DC2,A.DG3,A.DG4,A.DA5,A.DG6,A.DG7 2 type=lateral helix=#1 nts=7 GCGGAGG B.DG1,B.DC2,B.DG3,B.DG4,B.DA5,B.DG6,B.DG7