Detailed DSSR results for the G-quadruplex: PDB entry 6xrq
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 6xrq
- Class
- RNA
- Method
- X-ray (1.21 Å)
- Summary
- Structural descriptions of ligand interactions to DNA and RNA quadruplexes folded from the non-coding region of pseudorabies virus
- Reference
- Zhang Y, Bux K, Attana F, Wei D, Haider S, Parkinson GN (2024): "Structural descriptions of ligand interactions to RNA quadruplexes folded from the non-coding region of Pseudorabies virus." Biochimie. doi: 10.1016/j.biochi.2024.06.003.
- Abstract
- To rationalise the binding of specific ligands to RNA-quadruplex we investigated several naphthalene diimide ligands that interact with the non-coding region of Pseudorabies virus (PRV). Herein we report on the x-ray structure of the naphthalene diimide ND11 with an RNA G-quadruplex putative forming sequence from rPRV. Consistent with previously observed rPRV sequence it assembles into a bimolecular RNA G-quadruplex consisting of a pair of two tetrads stacked 3' to 5'. We observe that ND11 interacts by binding on both the externally available 5' and 3' quartets. The CUC (loop 1) is structurally altered to enhance the 5' mode of interaction. These loop residues are shifted significantly to generate a new ligand binding pocket whereas the terminal A14 residue is lifted away from the RNA G-quadruplex tetrad plane to be restacked above the bound ND11 ligand NDI core. CD analysis of this family of NDI ligands shows consistency in the spectra between the different ligands in the presence of the rPRV RNA G-quadruplex motif, reflecting a common folded topology and mode of ligand interaction. FRET melt assay confirms the strong stabilising properties of the tetrasubstituted NDI compounds and the contributions length of the substituted groups have on melt temperatures.
- G4 notes
- 4 G-tetrads, 1 G4 helix, 2 G4 stems, 1 G4 coaxial stack, parallel(4+0), UUUU, coaxial interfaces: 3'/5'
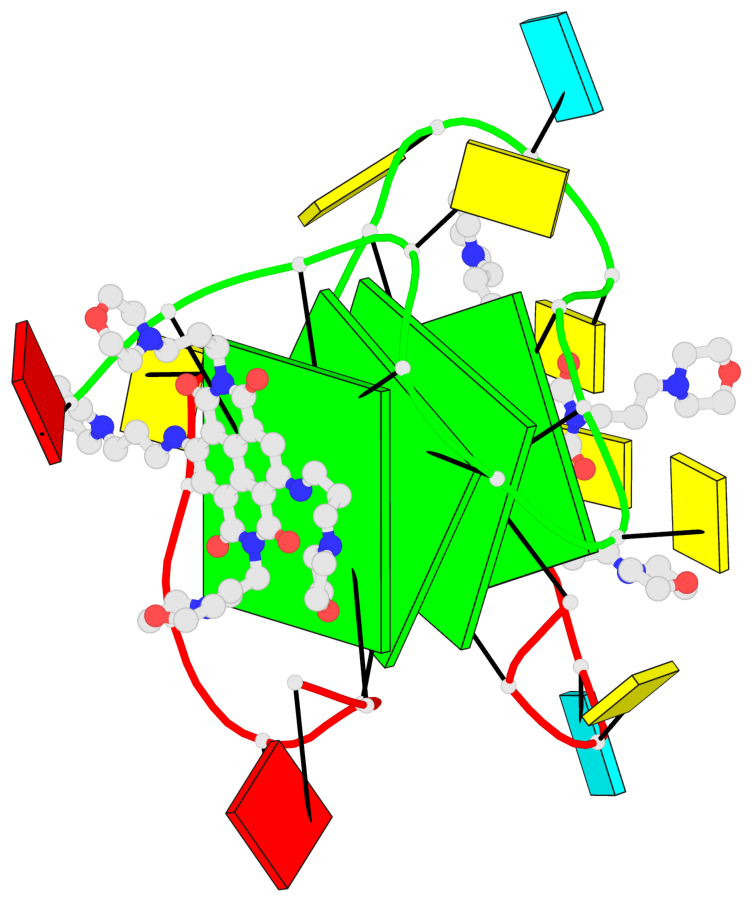
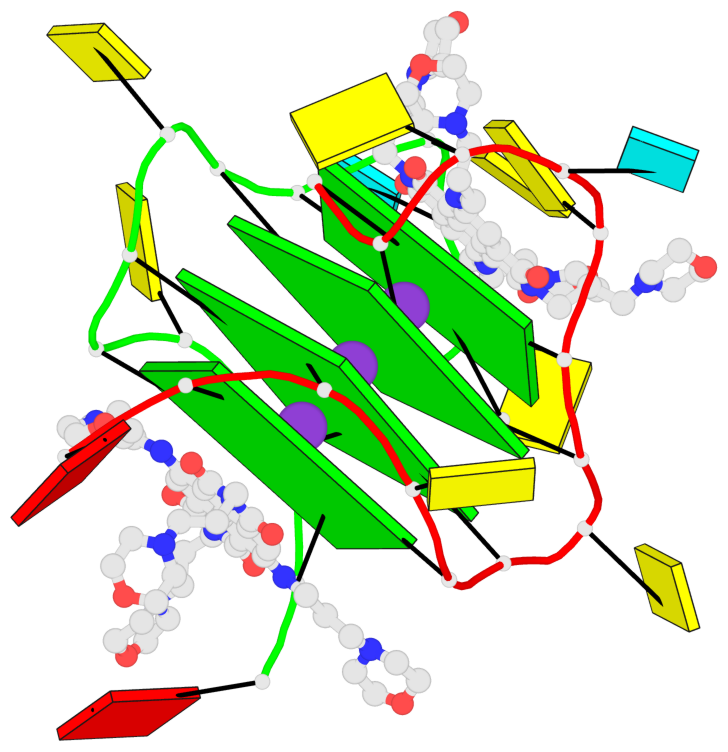
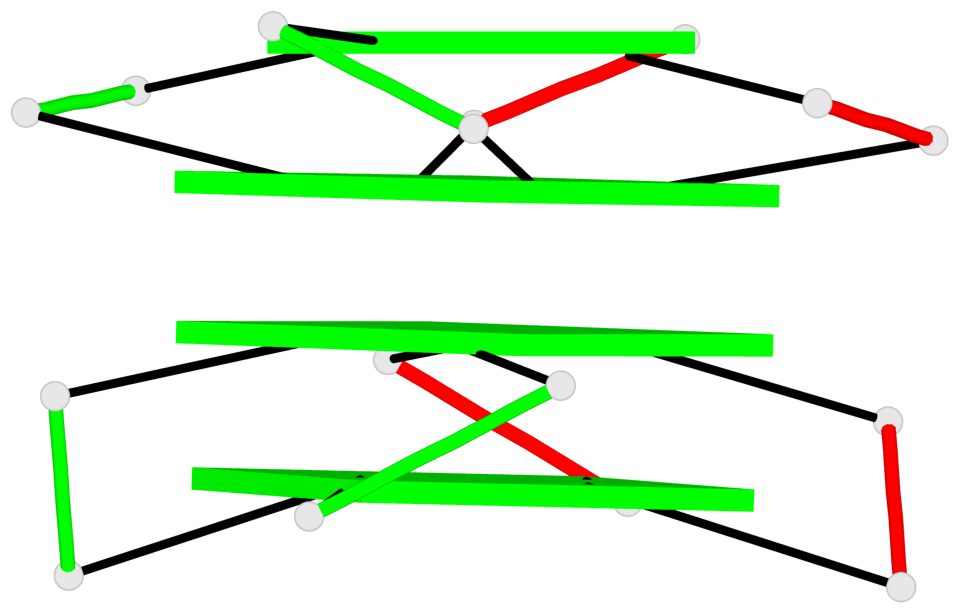
Base-block schematics in six views
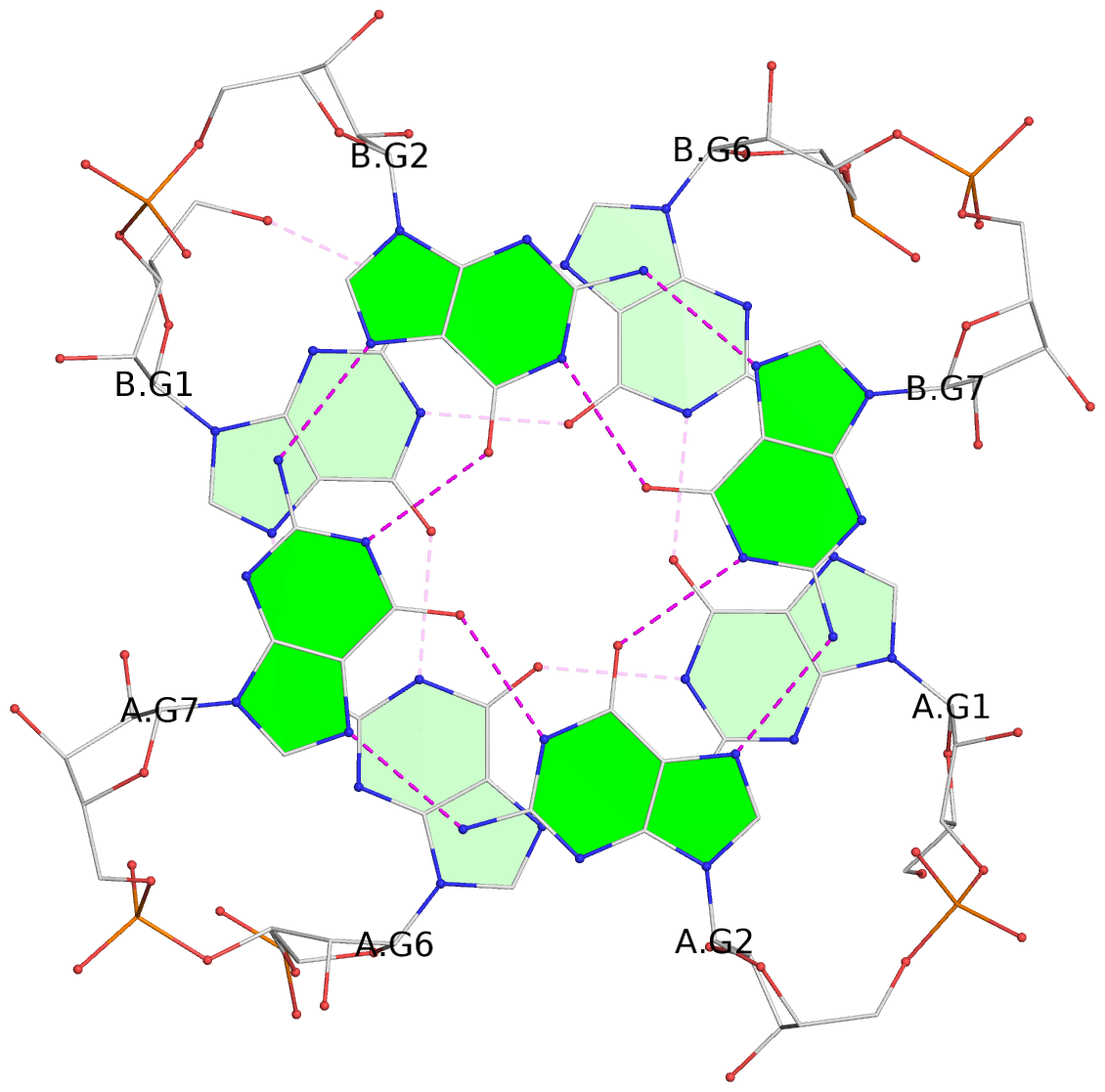
List of 4 G-tetrads
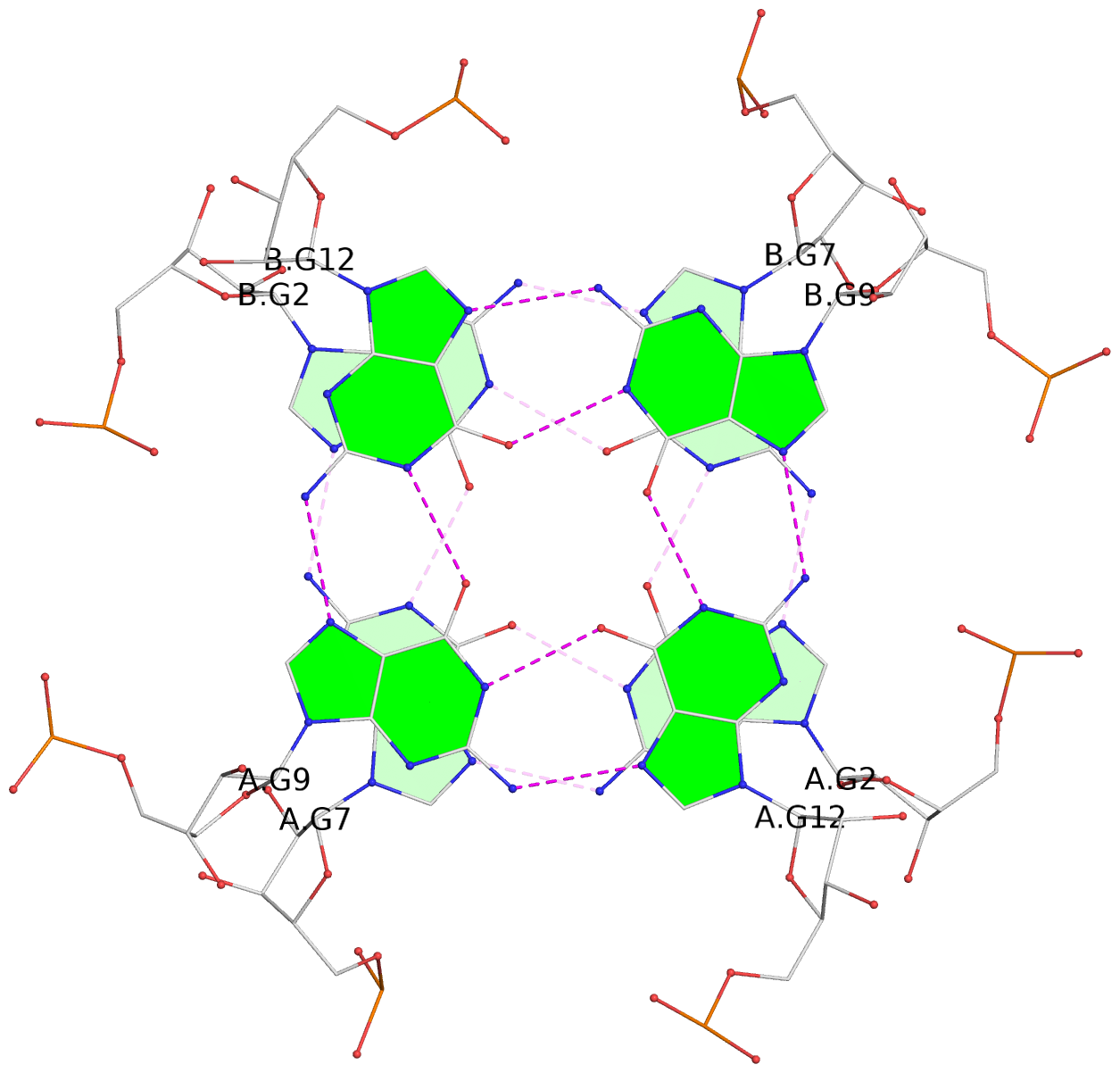
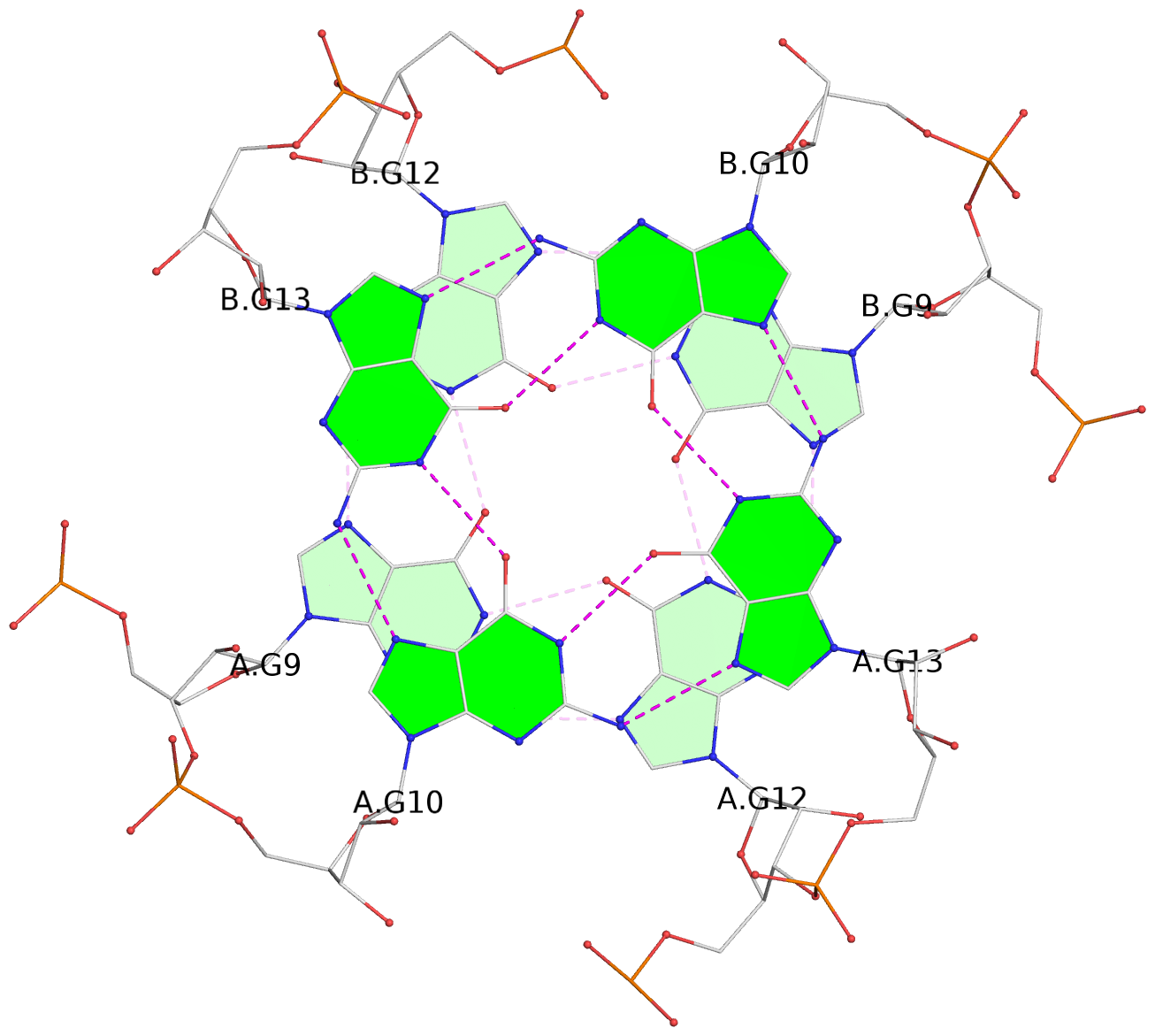
1 glyco-bond=ssss sugar=3333 groove=---- planarity=0.179 type=other nts=4 GGGG B.G1,A.G6,A.G1,B.G6 2 glyco-bond=---- sugar=---- groove=---- planarity=0.143 type=planar nts=4 GGGG B.G2,B.G7,A.G2,A.G7 3 glyco-bond=---- sugar=-3-3 groove=---- planarity=0.092 type=planar nts=4 GGGG B.G9,B.G12,A.G9,A.G12 4 glyco-bond=---- sugar=---3 groove=---- planarity=0.226 type=bowl-2 nts=4 GGGG B.G10,B.G13,A.G10,A.G13
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 4 G-tetrad layers, inter-molecular, with 2 stems
List of 2 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 2 G-tetrad layers, 2 loops, inter-molecular, UUUU, parallel, parallel(4+0)
Stem#2, 2 G-tetrad layers, 2 loops, inter-molecular, UUUU, parallel, parallel(4+0)
List of 1 G4 coaxial stack
1 G4 helix#1 contains 2 G4 stems: [#1,#2] [3'/5']