Detailed DSSR results for the G-quadruplex: PDB entry 6yep
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 6yep
- Class
- DNA
- Method
- NMR
- Summary
- Lna modified g-quadruplex with flipped g-tract and central tetrad
- Reference
- Haase L, Weisz K (2020): "Locked nucleic acid building blocks as versatile tools for advanced G-quadruplex design." Nucleic Acids Res., 48, 10555-10566. doi: 10.1093/nar/gkaa720.
- Abstract
- A hybrid-type G-quadruplex is modified with LNA (locked nucleic acid) and 2'-F-riboguanosine in various combinations at the two syn positions of its third antiparallel G-tract. LNA substitution in the central tetrad causes a complete rearrangement to either a V-loop or antiparallel structure, depending on further modifications at the 5'-neighboring site. In the two distinct structural contexts, LNA-induced stabilization is most effective compared to modifications with other G surrogates, highlighting a potential use of LNA residues for designing not only parallel but various more complex G4 structures. For instance, the conventional V-loop is a structural element strongly favored by an LNA modification at the V-loop 3'-end in contrast with an alternative V-loop, clearly distinguishable by altered conformational properties and base-backbone interactions as shown in a detailed analysis of V-loop structures.
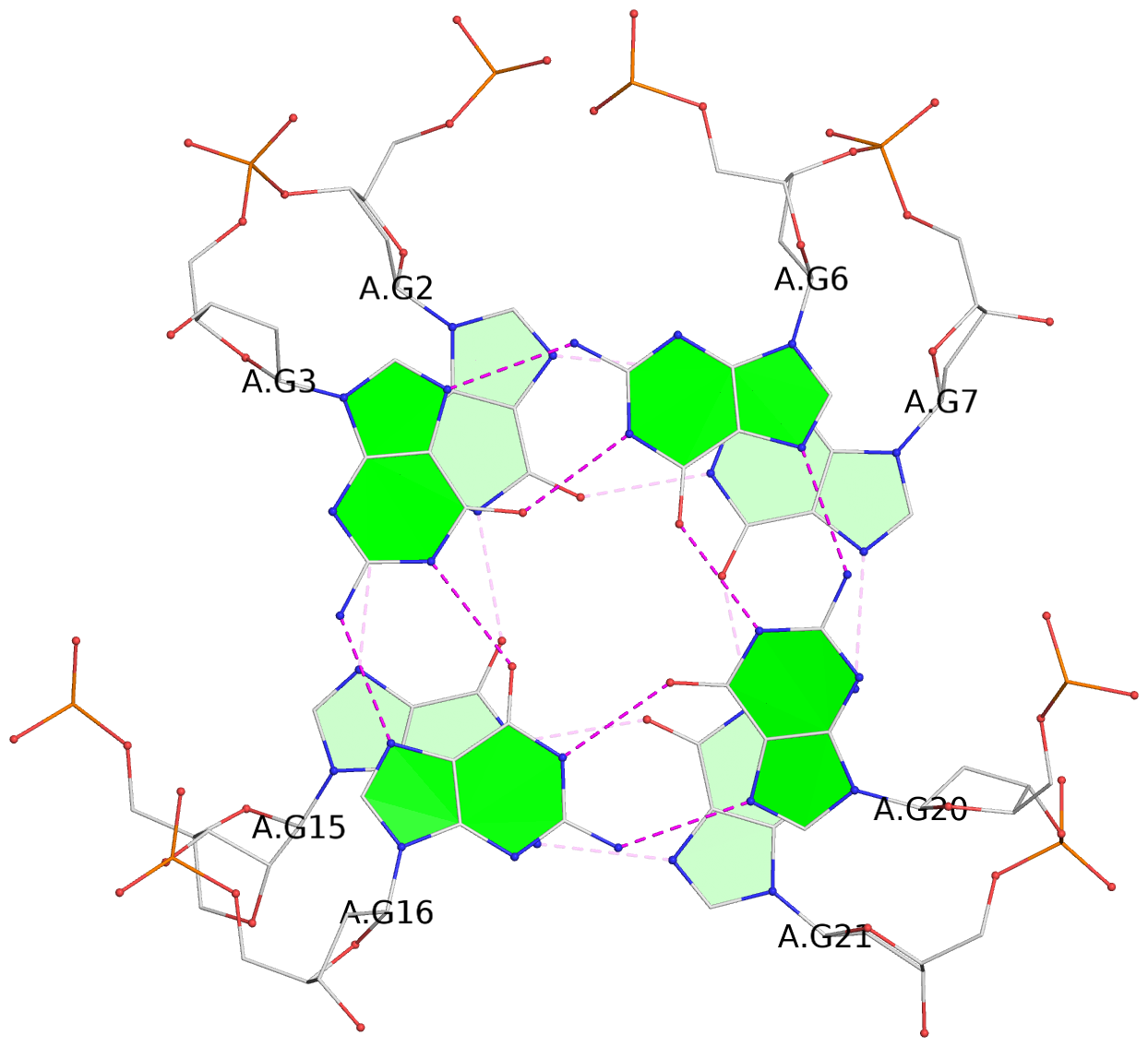
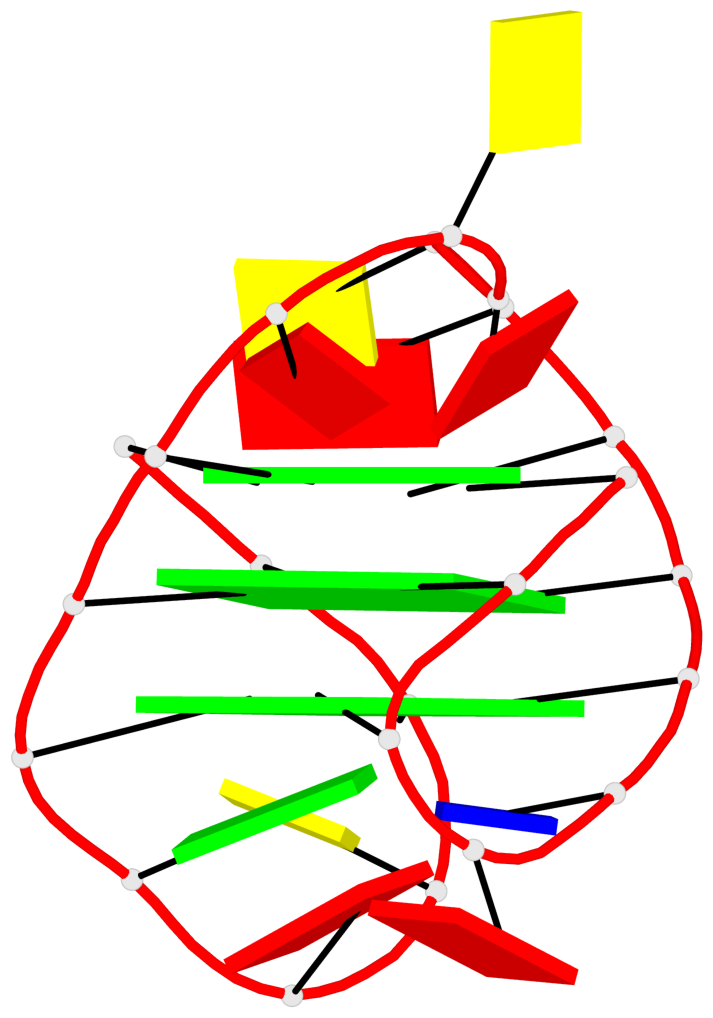
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(+LnD-Lw), basket(2+2), UUDD
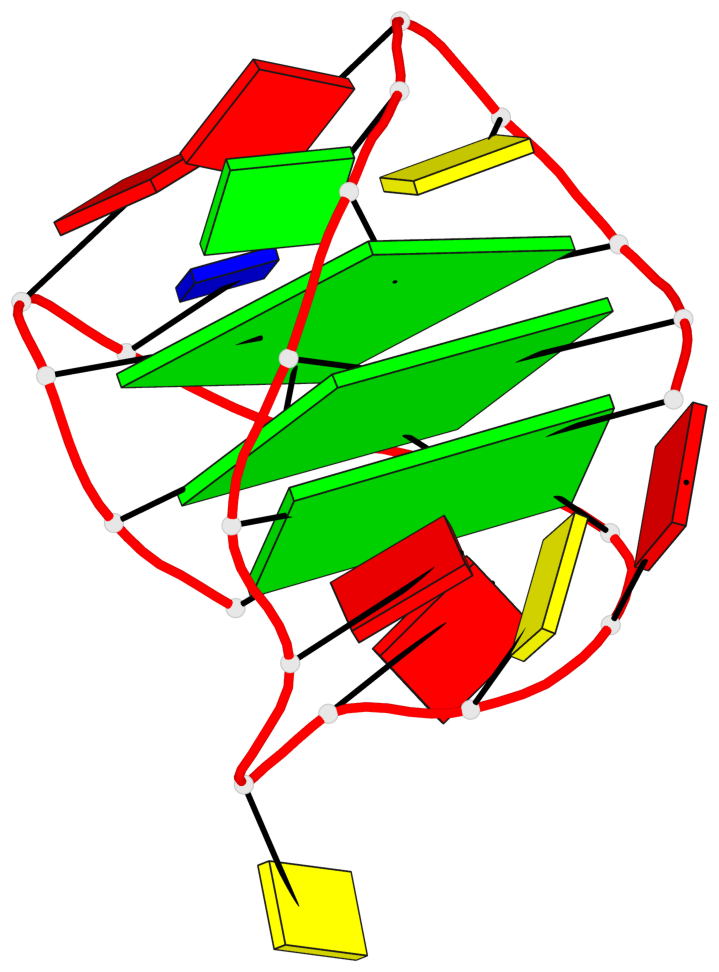
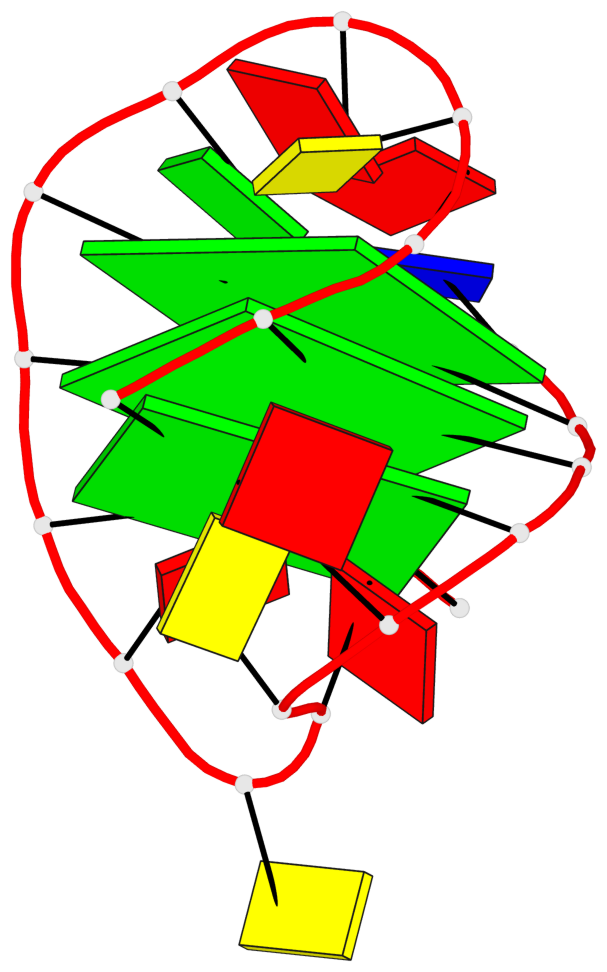
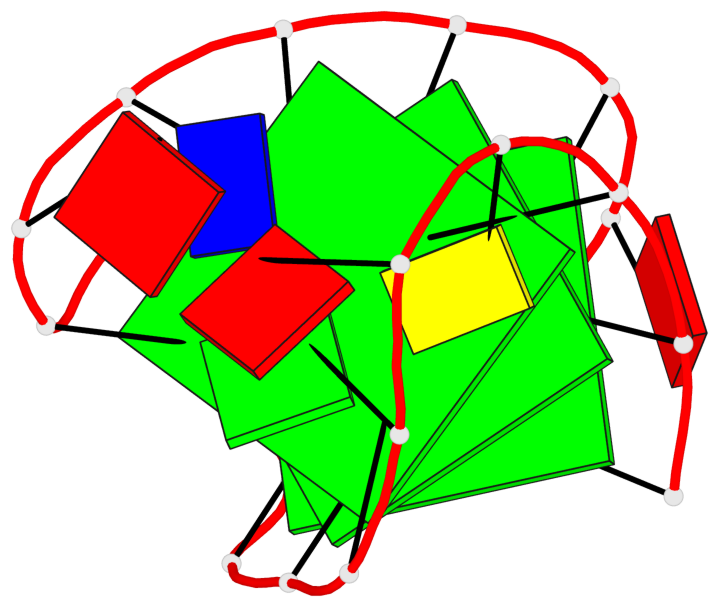
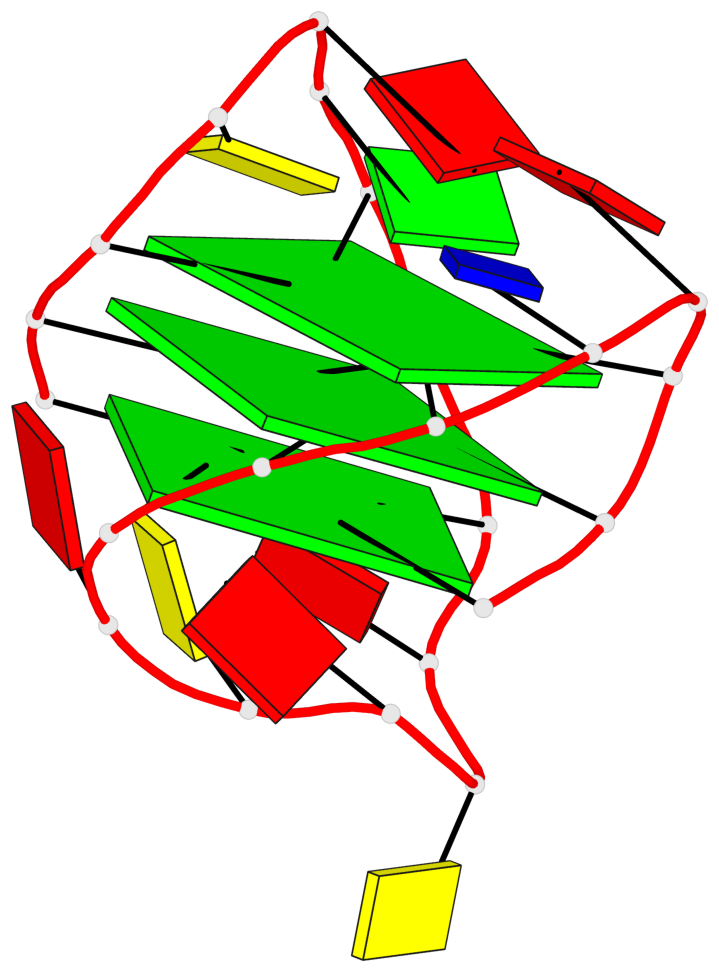
Base-block schematics in six views
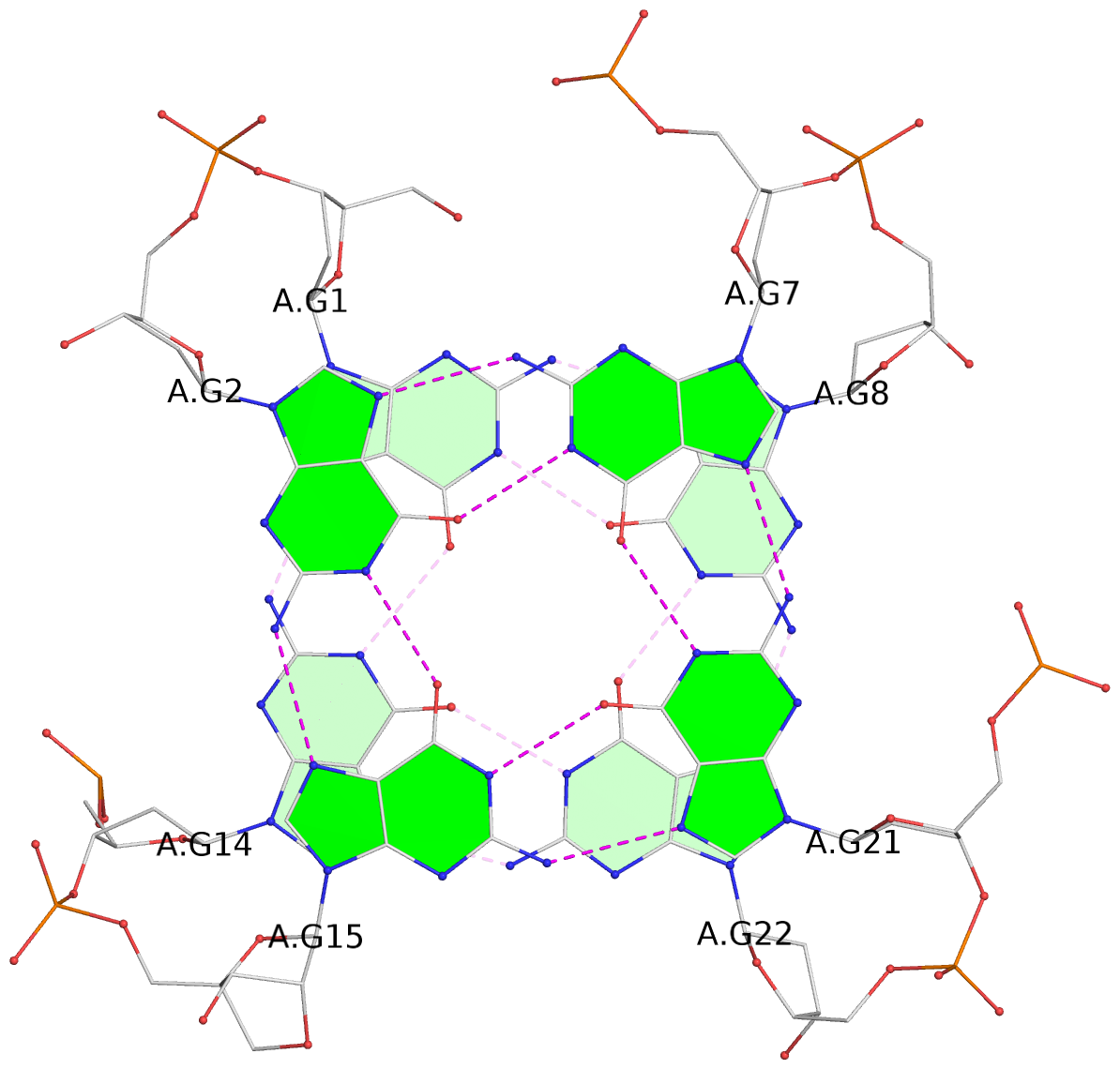
List of 3 G-tetrads
1 glyco-bond=ss-- sugar=---- groove=-w-n planarity=0.293 type=bowl nts=4 GGGG A.DG1,A.DG14,A.DG22,A.DG8 2 glyco-bond=--ss sugar=-3-- groove=-w-n planarity=0.193 type=other nts=4 GgGG A.DG2,A.LCG15,A.DG21,A.DG7 3 glyco-bond=--ss sugar=---- groove=-w-n planarity=0.238 type=other nts=4 GGGG A.DG3,A.DG16,A.DG20,A.DG6
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.