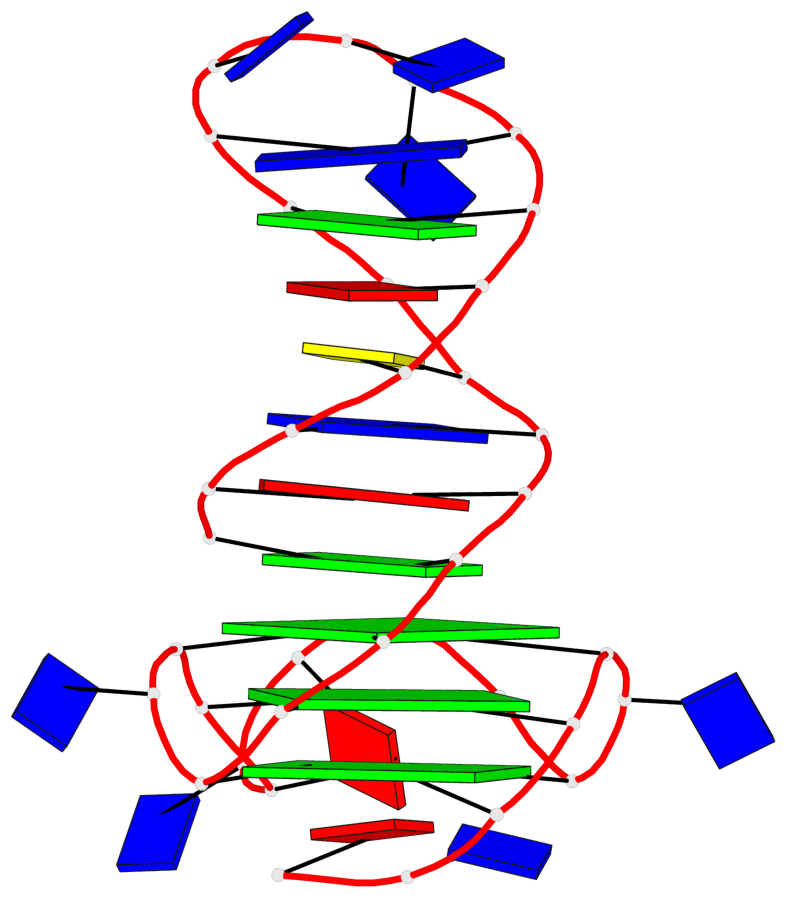
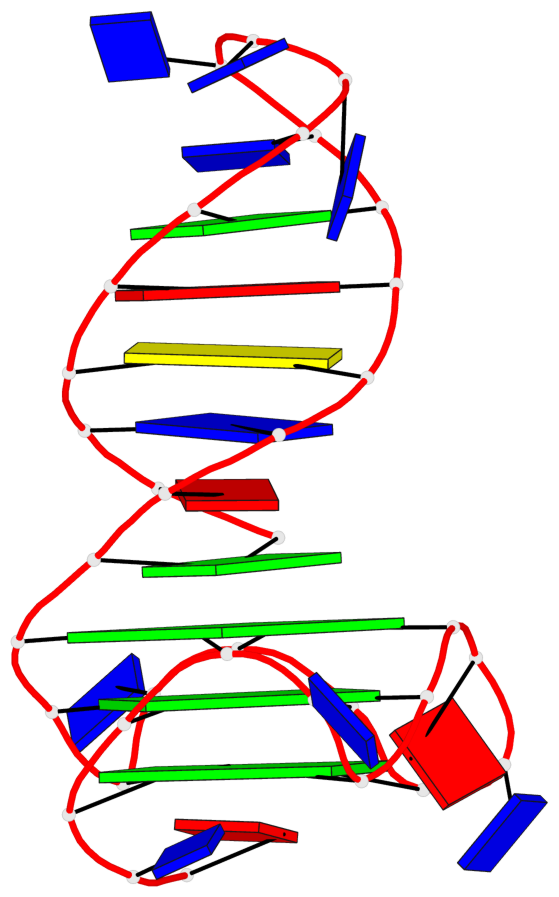
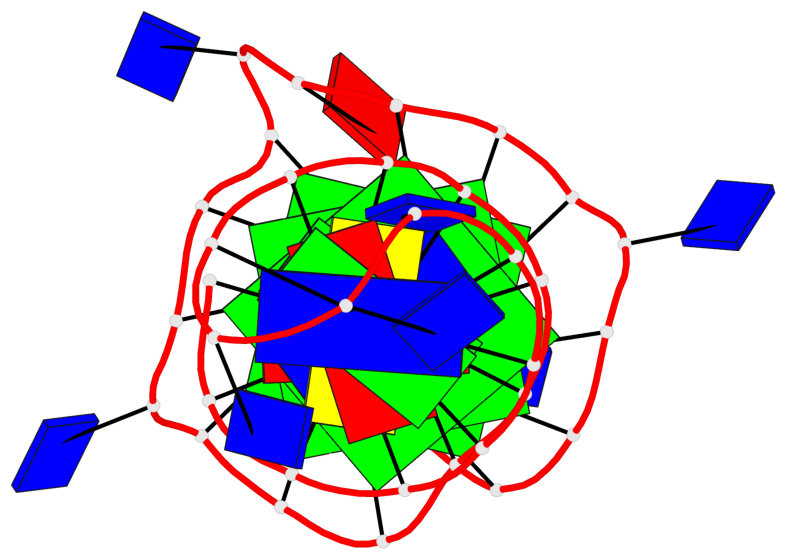
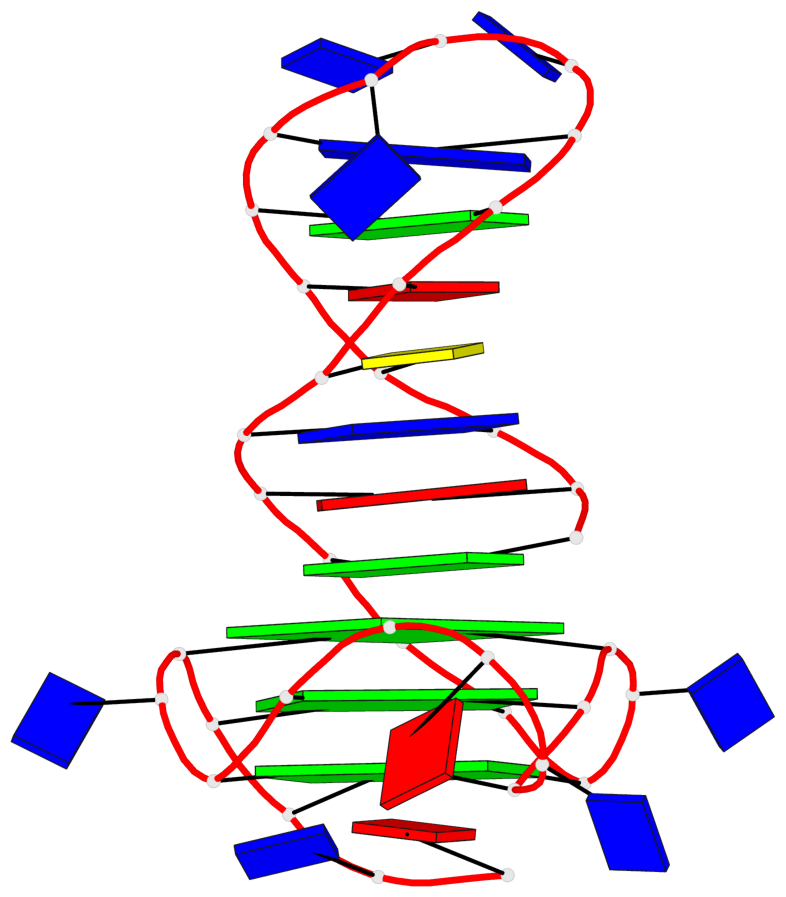
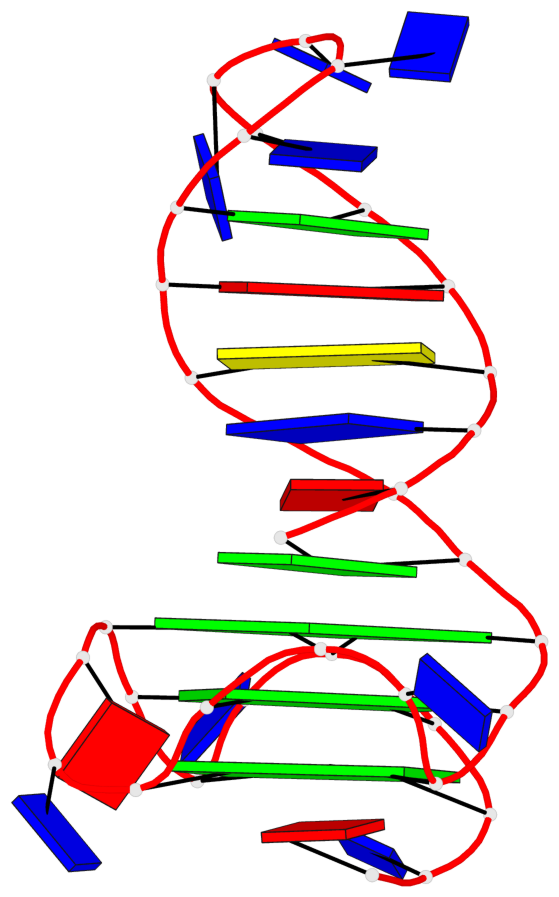
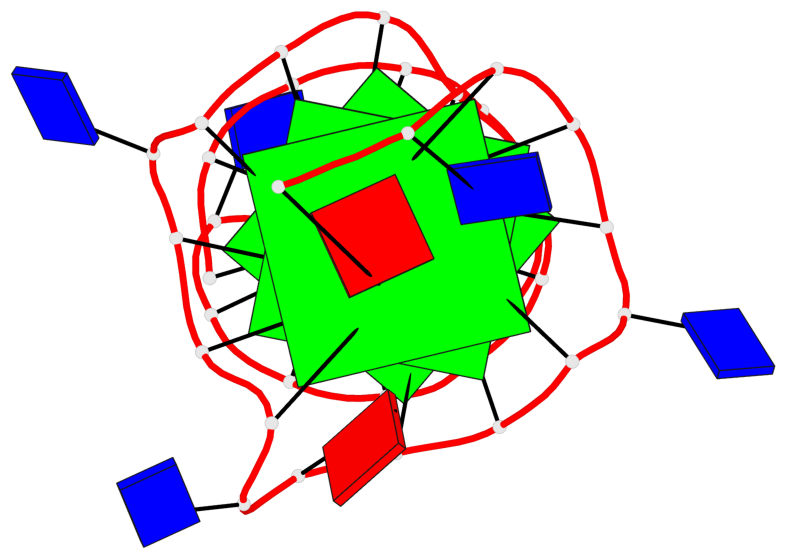
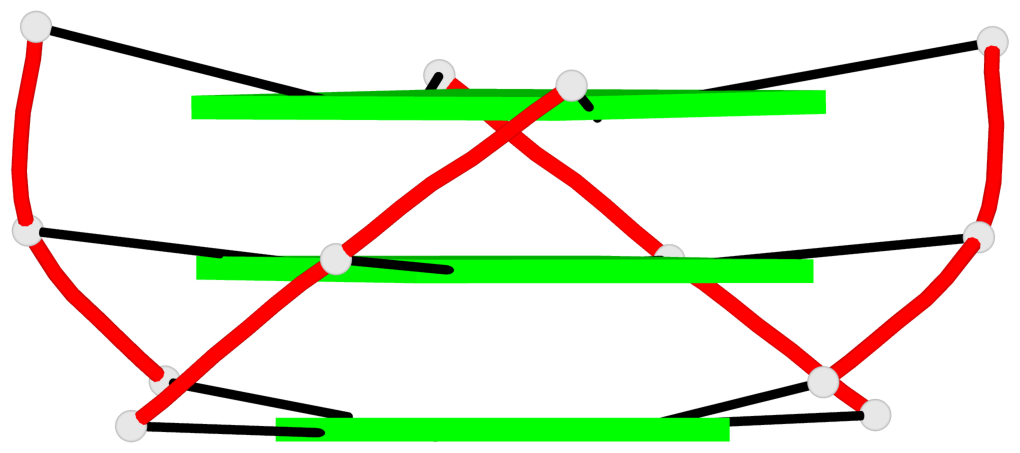
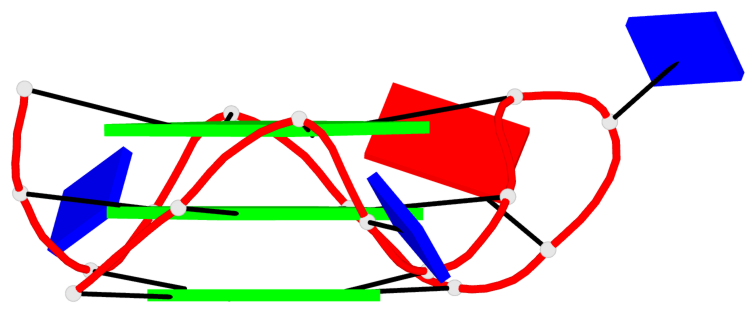
Detailed DSSR results for the G-quadruplex: PDB entry 6zl9
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 6zl9
- Class
- DNA
- Method
- NMR
- Summary
- Structure of a parallel c-myc modified with 5' duplex stem-loop overhang
- Reference
- Vianney YM, Preckwinkel P, Mohr S, Weisz K (2020): "Quadruplex-Duplex Junction: A High-Affinity Binding Site for Indoloquinoline Ligands." Chemistry, 26, 16910-16922. doi: 10.1002/chem.202003540.
- Abstract
- A parallel quadruplex derived from the Myc promoter sequence was extended by a stem-loop duplex at either its 5'- or 3'-terminus to mimic a quadruplex-duplex (Q-D) junction as a potential genomic target. High-resolution structures of the hybrids demonstrate continuous stacking of the duplex on the quadruplex core without significant perturbations. An indoloquinoline ligand carrying an aminoalkyl side chain was shown to bind the Q-D hybrids with a very high affinity in the order Ka ≈107 m-1 irrespective of the duplex location at the quadruplex 3'- or 5'-end. NMR chemical shift perturbations identified the tetrad face of the Q-D junction as specific binding site for the ligand. However, calorimetric analyses revealed significant differences in the thermodynamic profiles upon binding to hybrids with either a duplex extension at the quadruplex 3'- or 5'-terminus. A large enthalpic gain and considerable hydrophobic effects are accompanied by the binding of one ligand to the 3'-Q-D junction, whereas non-hydrophobic entropic contributions favor binding with formation of a 2:1 ligand-quadruplex complex in case of the 5'-Q-D hybrid.
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-P-P-P), parallel(4+0), UUUU
Base-block schematics in six views
List of 3 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.188 type=other nts=4 GGGG A.DG18,A.DG22,A.DG27,A.DG31 2 glyco-bond=---- sugar=---- groove=---- planarity=0.227 type=other nts=4 GGGG A.DG19,A.DG23,A.DG28,A.DG32 3 glyco-bond=---- sugar=---- groove=---- planarity=0.213 type=other nts=4 GGGG A.DG20,A.DG24,A.DG29,A.DG33
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.