Detailed DSSR results for the G-quadruplex: PDB entry 7cv3
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 7cv3
- Class
- DNA
- Method
- NMR
- Summary
- Quadruplex-duplex hybrid structure in the pim1 gene, form 1
- Reference
- Tan DJY, Winnerdy FR, Lim KW, Phan AT (2020): "Coexistence of two quadruplex-duplex hybrids in the PIM1 gene." Nucleic Acids Res., 48, 11162-11171. doi: 10.1093/nar/gkaa752.
- Abstract
- The triple-negative breast cancer (TNBC), a subtype of breast cancer which lacks of targeted therapies, exhibits a poor prognosis. It was shown recently that the PIM1 oncogene is highly related to the proliferation of TNBC cells. A quadruplex-duplex hybrid (QDH) forming sequence was recently found to exist near the transcription start site of PIM1. This structure could be an attractive target for regulation of the PIM1 gene expression and thus the treatment of TNBC. Here, we present the solution structures of two QDHs that could coexist in the human PIM1 gene. Form 1 is a three-G-tetrad-layered (3+1) G-quadruplex containing a propeller loop, a lateral loop and a stem-loop made up of three G•C Watson-Crick base pairs. On the other hand, Form 2 is an anti-parallel G-quadruplex comprising two G-tetrads and a G•C•G•C tetrad; the structure has three lateral loops with the middle stem-loop made up of two Watson-Crick G•C base pairs. These structures provide valuable information for the design of G-quadruplex-specific ligands for PIM1 transcription regulation.
- G4 notes
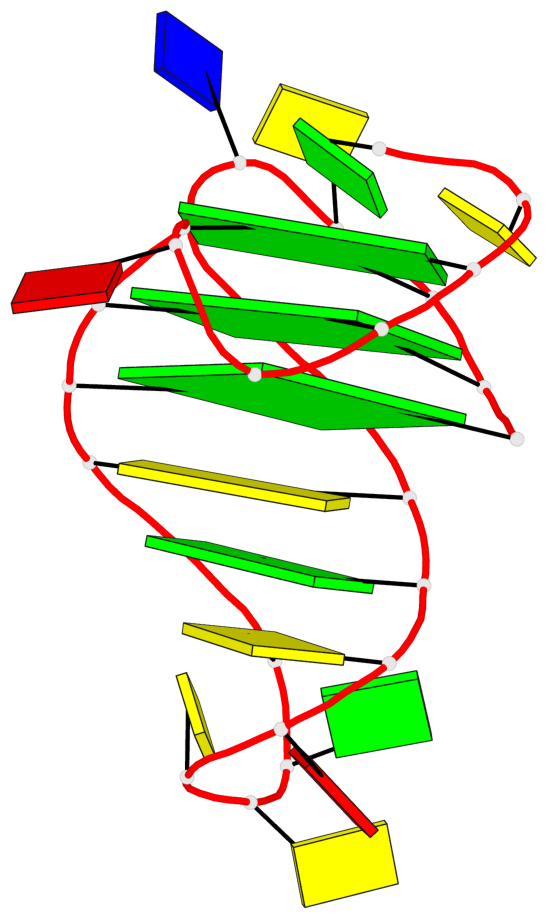
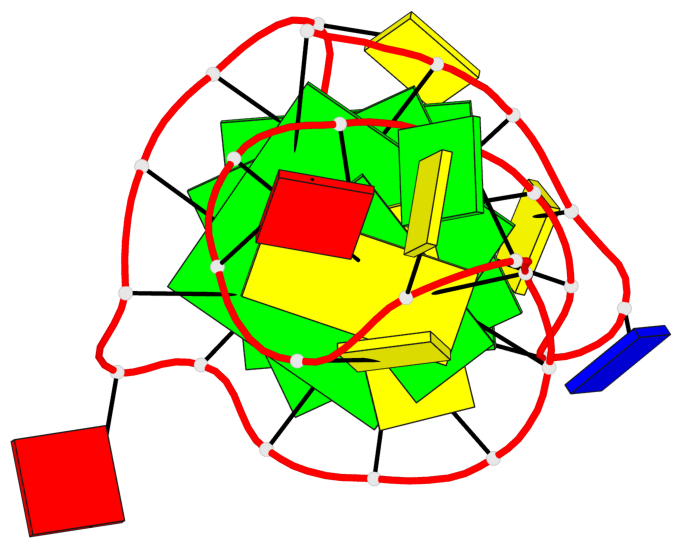
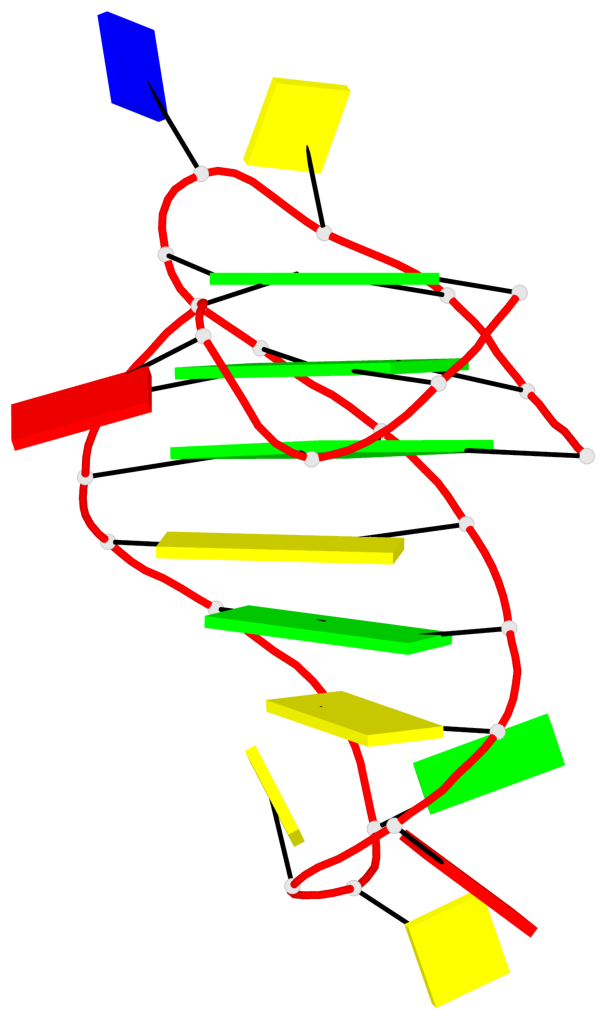
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-P-Lw-Ln), hybrid-1(3+1), UUDU
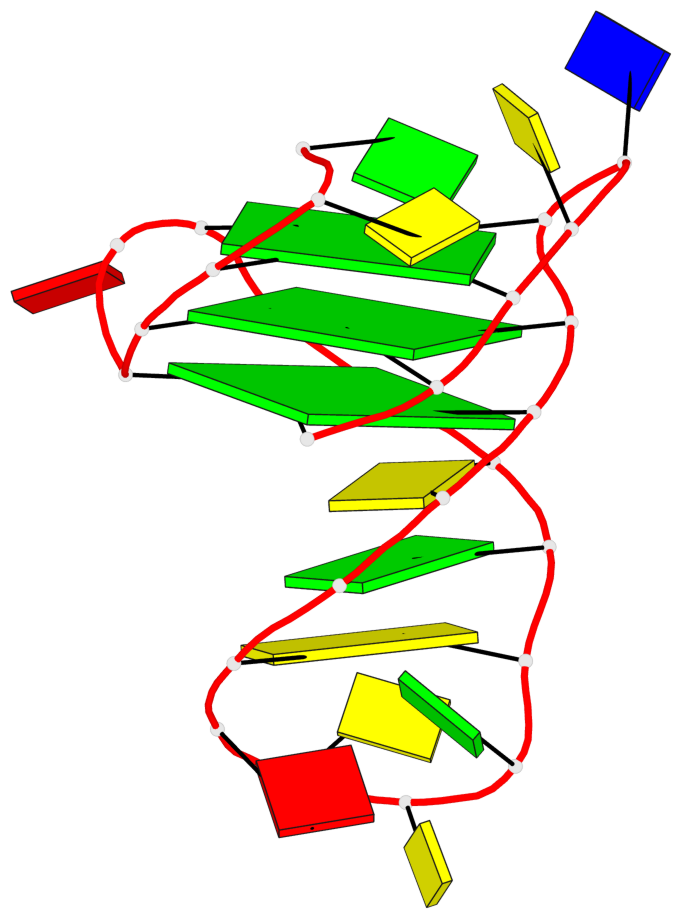
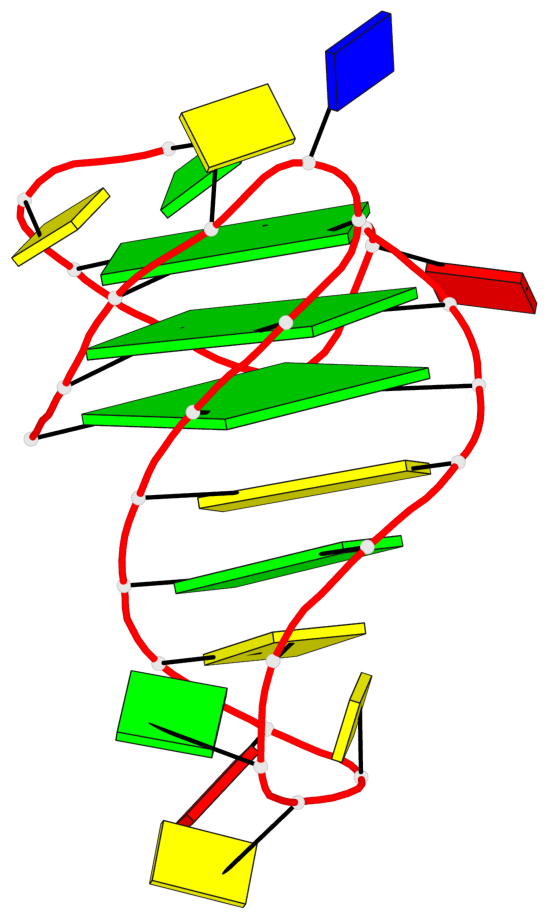
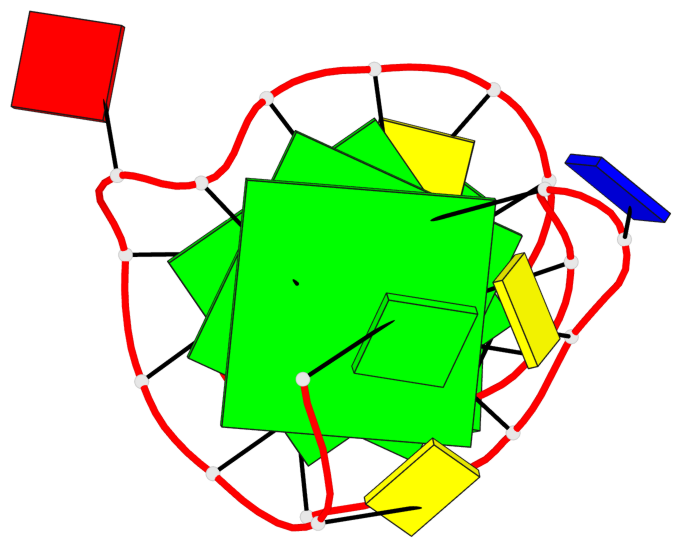
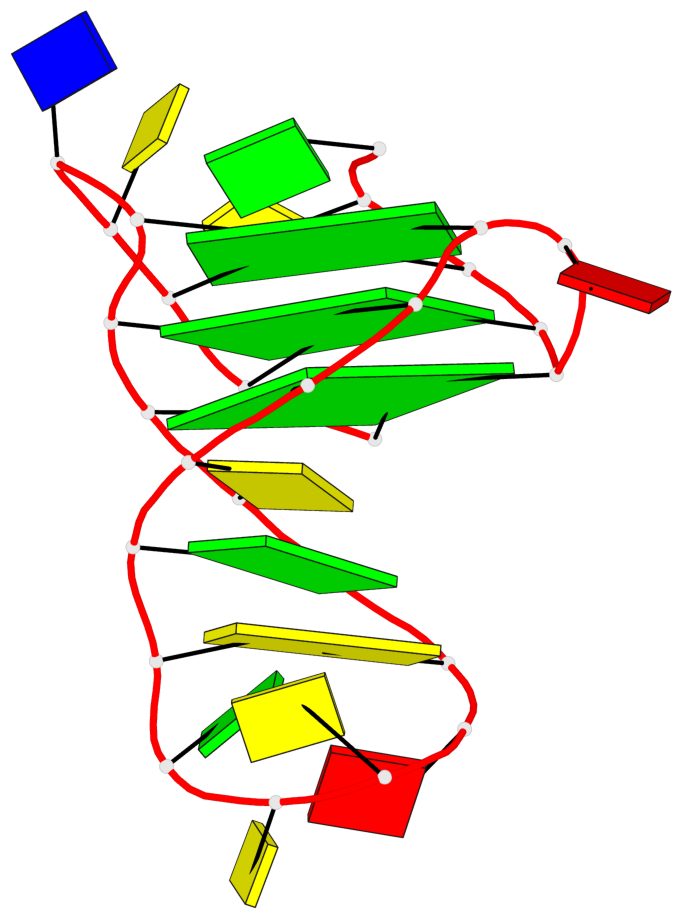
Base-block schematics in six views
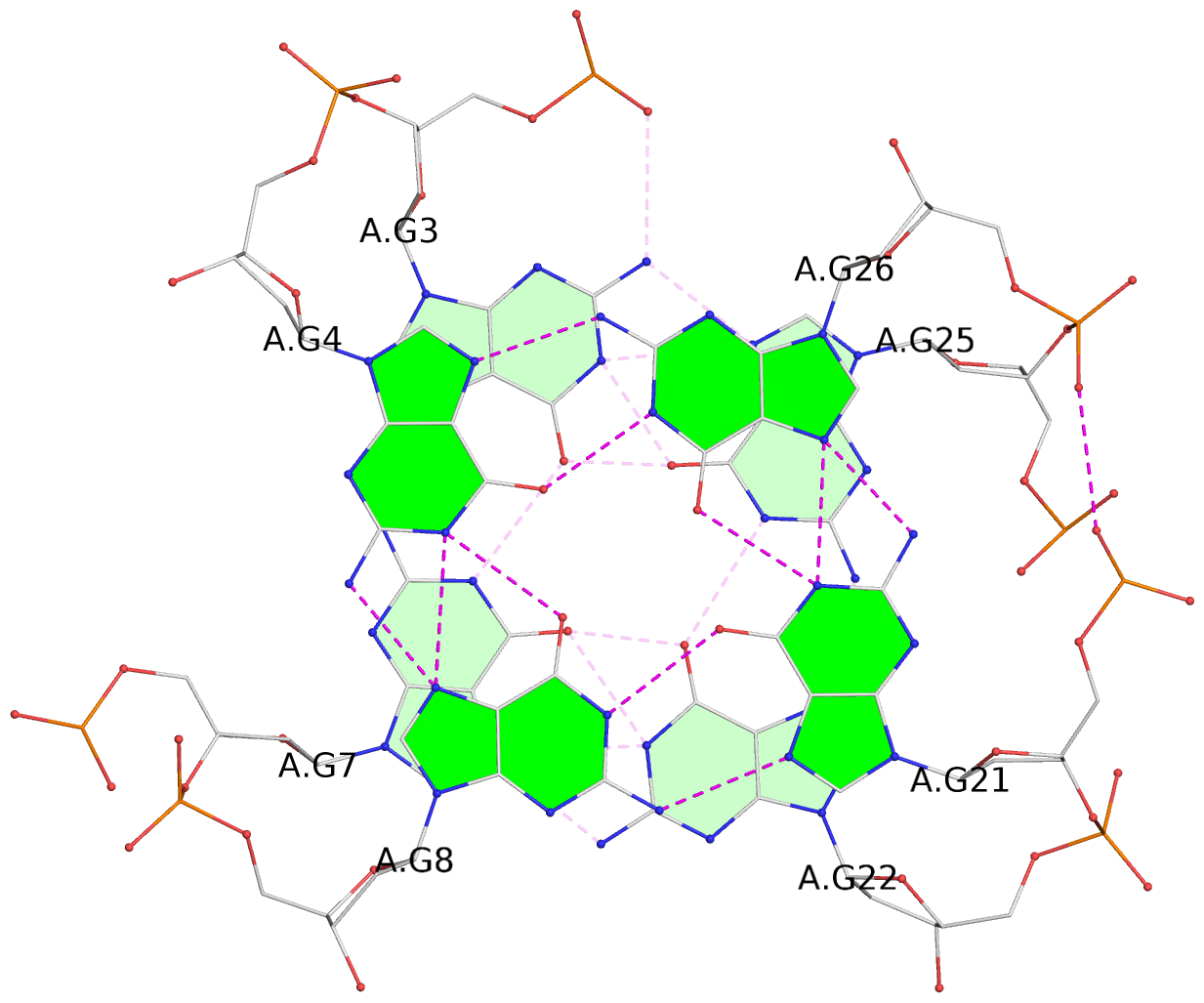
List of 3 G-tetrads
1 glyco-bond=ss-s sugar=--.- groove=-wn- planarity=0.095 type=planar nts=4 GGGG A.DG3,A.DG7,A.DG22,A.DG25 2 glyco-bond=--s- sugar=---- groove=-wn- planarity=0.101 type=planar nts=4 GGGG A.DG4,A.DG8,A.DG21,A.DG26 3 glyco-bond=--s- sugar=---- groove=-wn- planarity=0.063 type=planar nts=4 GGGG A.DG5,A.DG9,A.DG20,A.DG27
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.