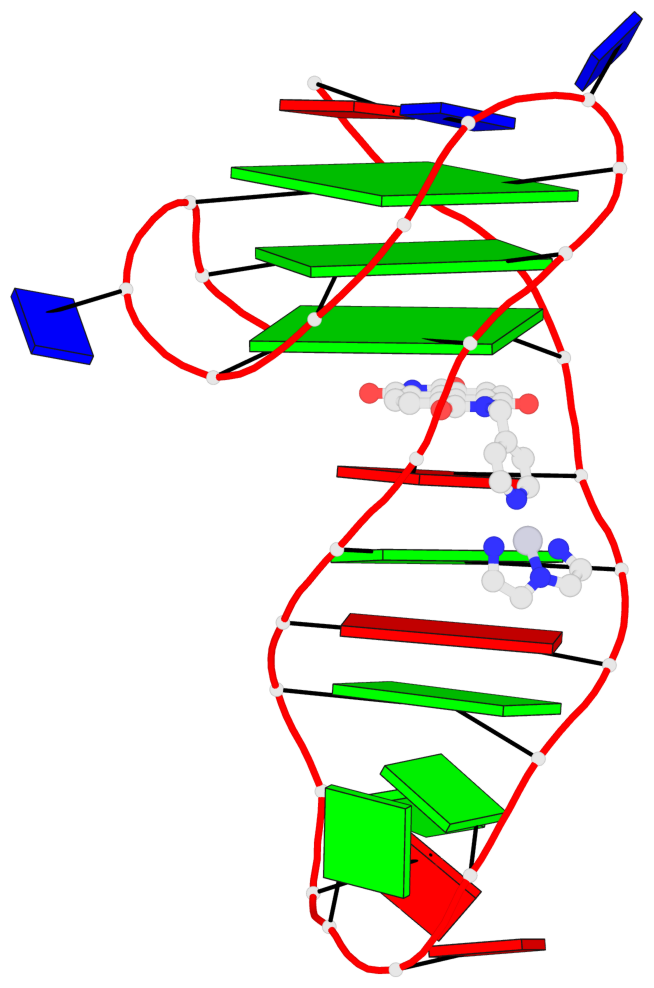
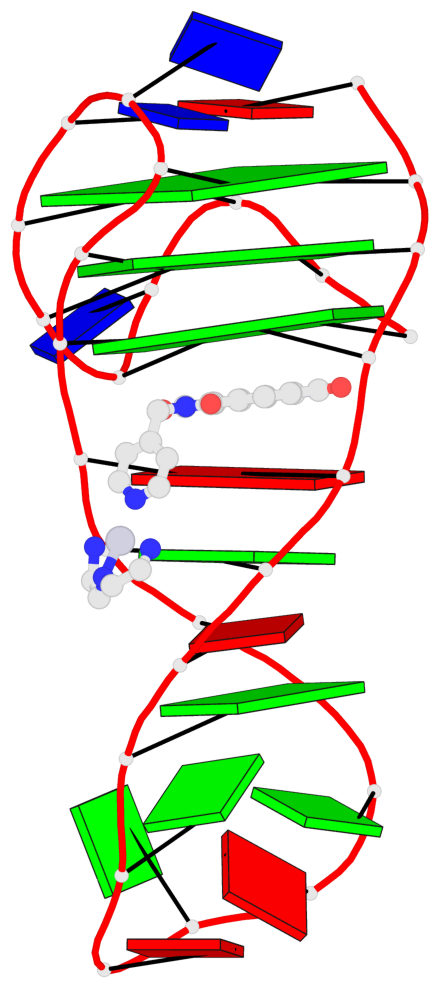
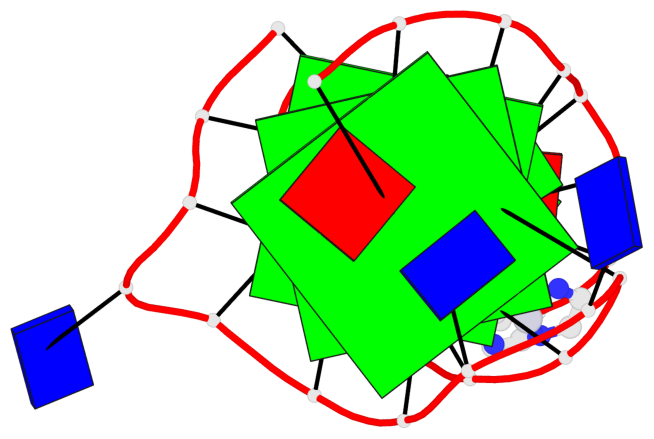
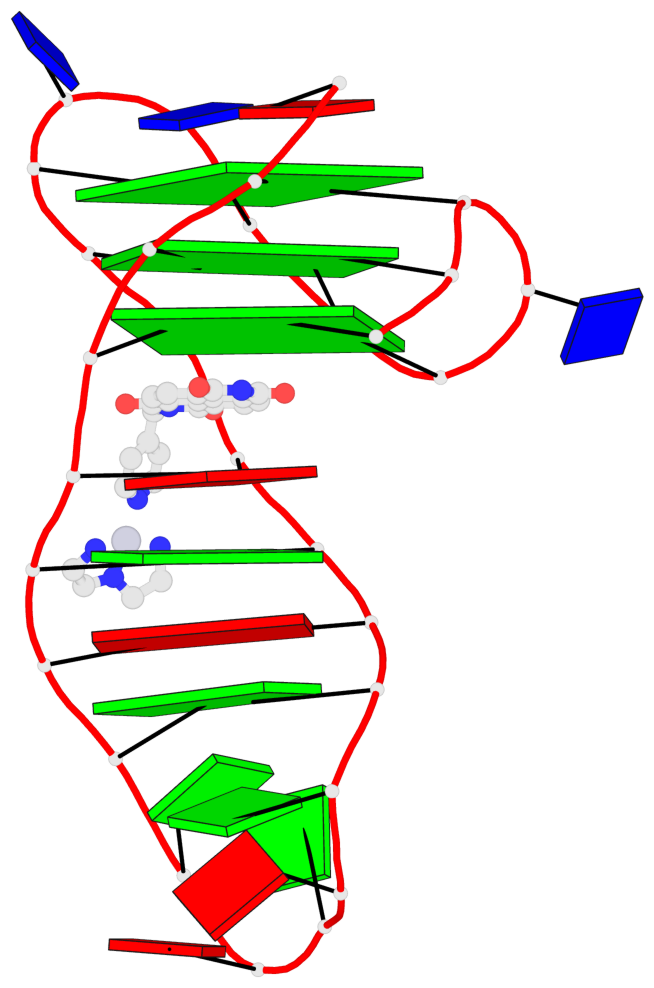
Detailed DSSR results for the G-quadruplex: PDB entry 7el7
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 7el7
- Class
- DNA
- Method
- NMR
- Summary
- NMR solution structure of the 1:1 complex of a quadruplex-duplex hybrid myt1l and a platinum(ii) ligand l1pt(dien)
- Reference
- Liu LY, Wang KN, Liu W, Zeng YL, Hou MX, Yang J, Mao ZW (2021): "Spatial Matching Selectivity and Solution Structure of Organic-Metal Hybrid to Quadruplex-Duplex Hybrid." Angew.Chem.Int.Ed.Engl., 60, 20833-20839. doi: 10.1002/anie.202106256.
- Abstract
- The sequence-dependent DNA secondary structures possess structure polymorphism. To date, studies on regulated ligands mainly focus on individual DNA secondary topologies, while lack focus on quadruplex-duplex hybrids (QDHs). Here, we design an organic-metal hybrid ligand L1 Pt(dien), which matches and selectively binds one type of QDHs with lateral duplex stem-loop (QLDH) with high affinity, while shows poor affinity for other QDHs and individual G4 or duplex DNA. The solution structure of QLDH MYT1L-L1 Pt(dien) complex was determined by NMR. The structure reveals that L1 Pt(dien) presents a chair-type conformation, whose large aromatic "chair surface" intercalates into the G-quadruplex-duplex interface via π-π stacking and "backrest" platinum unit interacts with duplex region through hydrogen bonding and electrostatic interactions, showing a highly matched lock-key binding mode. Our work provided guidance for spatial matching design of selectively targeting ligands to QDH structures.
- G4 notes
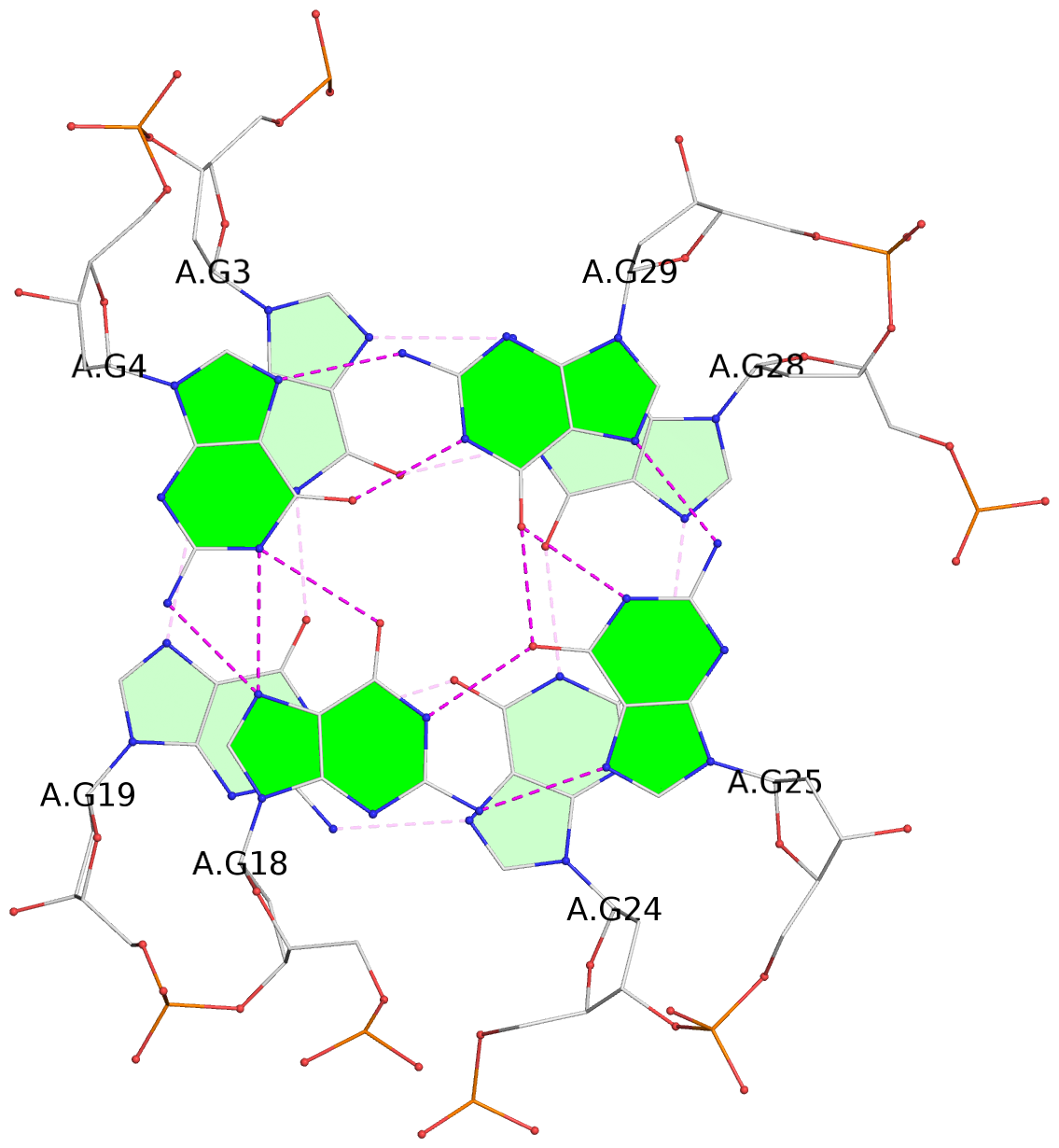
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-Lw-Ln-P), hybrid-2(3+1), UDUU
Base-block schematics in six views
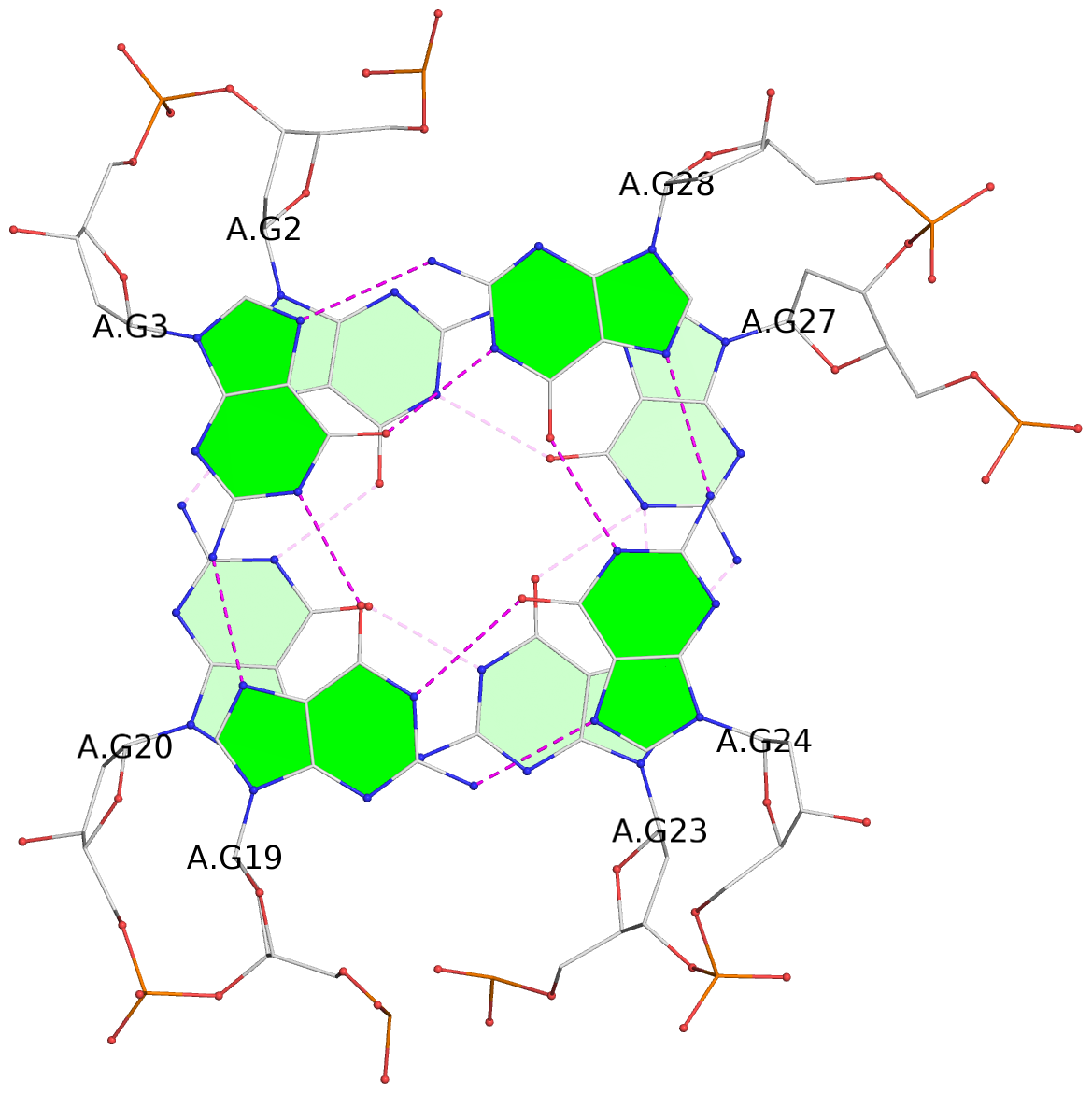
List of 3 G-tetrads
1 glyco-bond=s-ss sugar=-... groove=wn-- planarity=0.191 type=other nts=4 GGGG A.DG2,A.DG20,A.DG23,A.DG27 2 glyco-bond=-s-- sugar=.-.. groove=wn-- planarity=0.170 type=other nts=4 GGGG A.DG3,A.DG19,A.DG24,A.DG28 3 glyco-bond=-s-- sugar=.-.. groove=wn-- planarity=0.086 type=planar nts=4 GGGG A.DG4,A.DG18,A.DG25,A.DG29
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.