Detailed DSSR results for the G-quadruplex: PDB entry 7pnl
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 7pnl
- Class
- DNA
- Method
- X-ray (1.83 Å)
- Summary
- Complex between monomolecular human telomeric g-quadruplex and a sulfonamide derivative of the natural alkaloid berberine
- Reference
- Nocentini A, Di Porzio A, Bonardi A, Bazzicalupi C, Petreni A, Biver T, Bua S, Marzano S, Amato J, Pagano B, Iaccarino N, De Tito S, Amente S, Supuran CT, Randazzo A, Gratteri P (2024): "Development of a multi-targeted chemotherapeutic approach based on G-quadruplex stabilisation and carbonic anhydrase inhibition." J Enzyme Inhib Med Chem, 39, 2366236. doi: 10.1080/14756366.2024.2366236.
- Abstract
- A novel class of compounds designed to hit two anti-tumour targets, G-quadruplex structures and human carbonic anhydrases (hCAs) IX and XII is proposed. The induction/stabilisation of G-quadruplex structures by small molecules has emerged as an anticancer strategy, disrupting telomere maintenance and reducing oncogene expression. hCAs IX and XII are well-established anti-tumour targets, upregulated in many hypoxic tumours and contributing to metastasis. The ligands reported feature a berberine G-quadruplex stabiliser scaffold connected to a moiety inhibiting hCAs IX and XII. In vitro experiments showed that our compounds selectively stabilise G-quadruplex structures and inhibit hCAs IX and XII. The crystal structure of a telomeric G-quadruplex in complex with one of these ligands was obtained, shedding light on the ligand/target interaction mode. The most promising ligands showed significant cytotoxicity against CA IX-positive HeLa cancer cells in hypoxia, and the ability to stabilise G-quadruplexes within tumour cells.
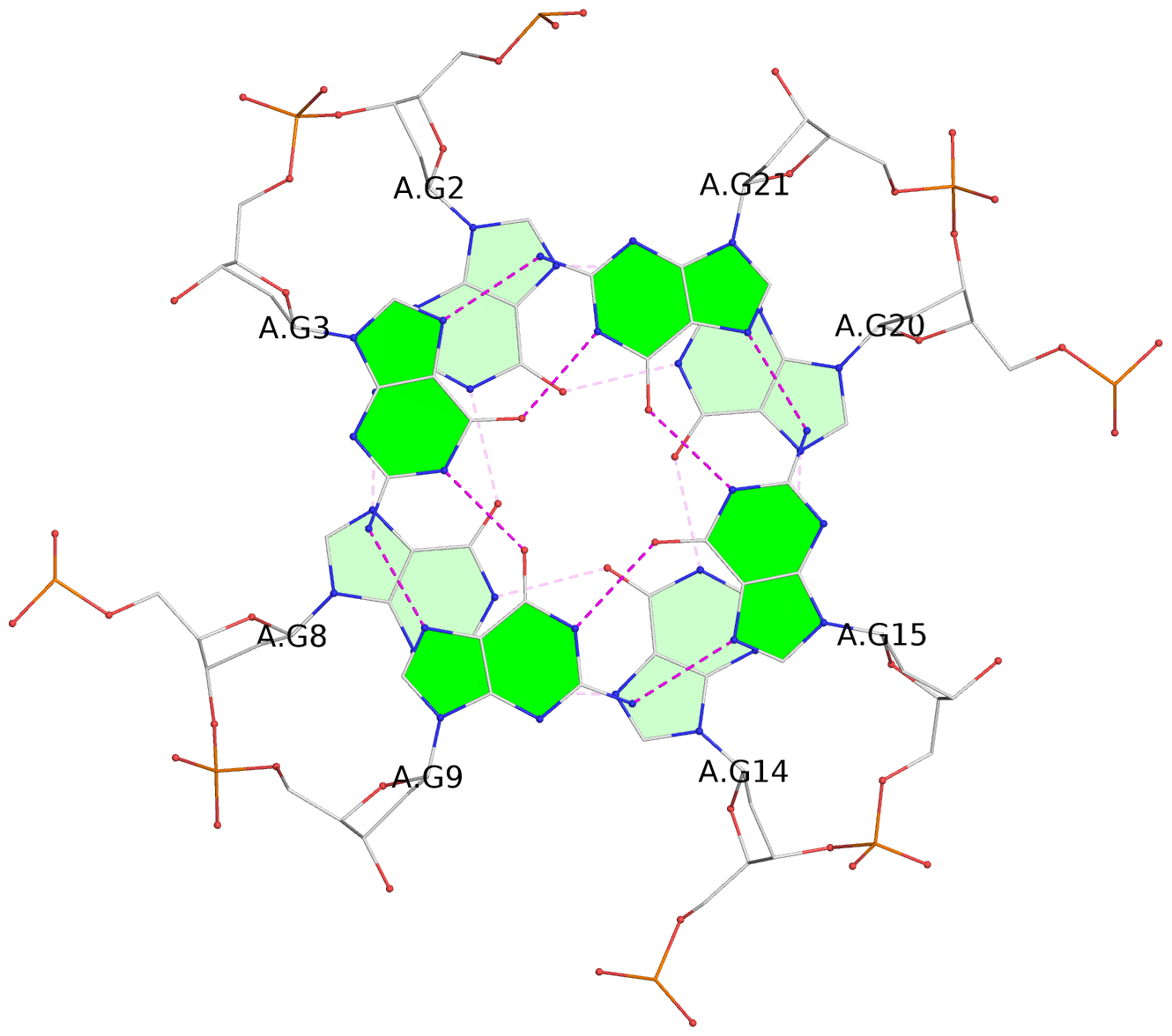
- G4 notes
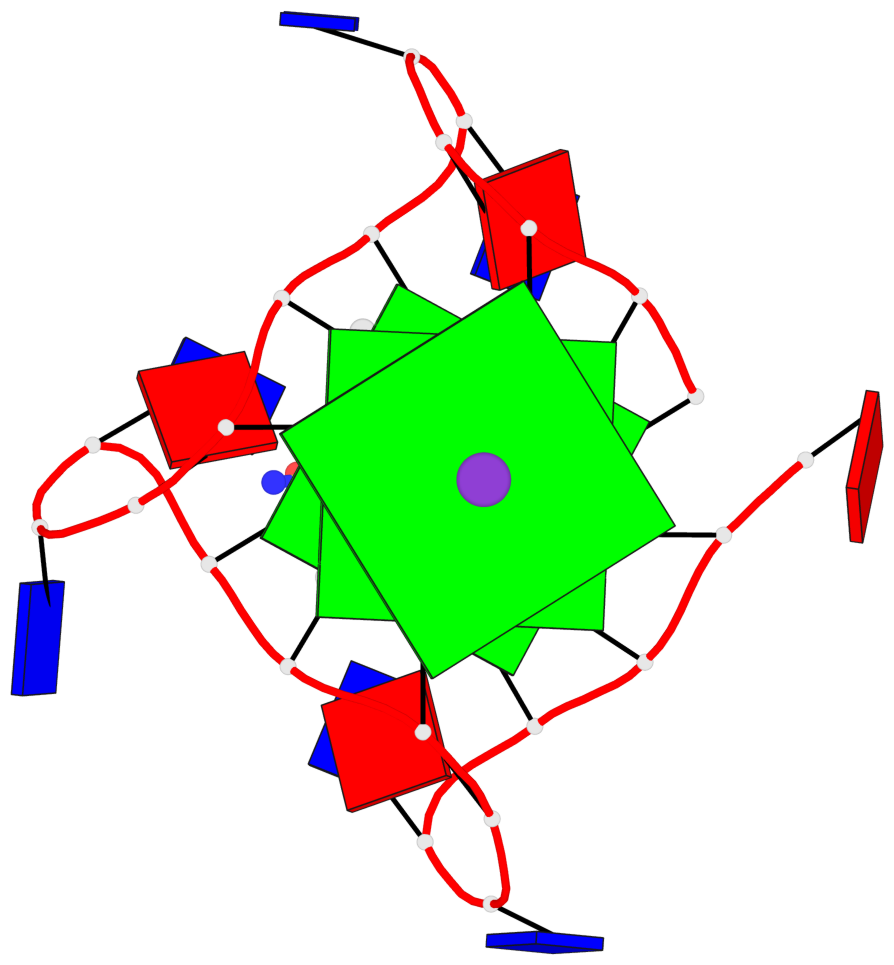
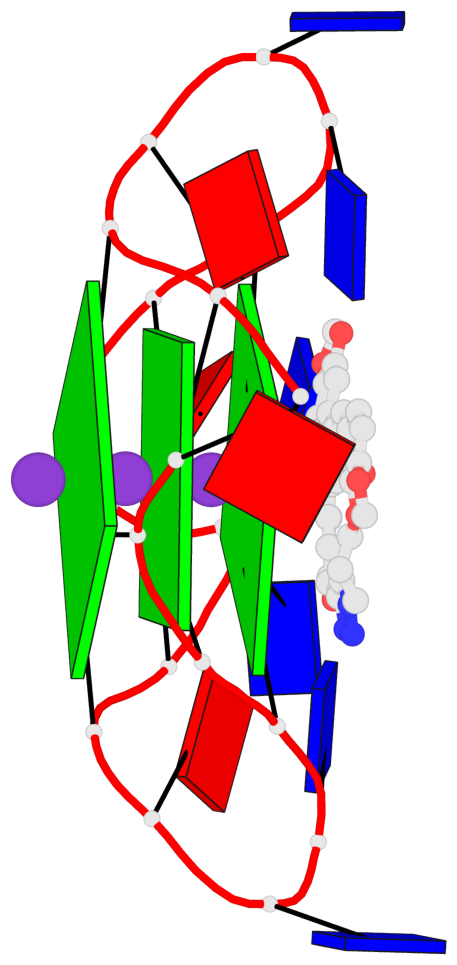
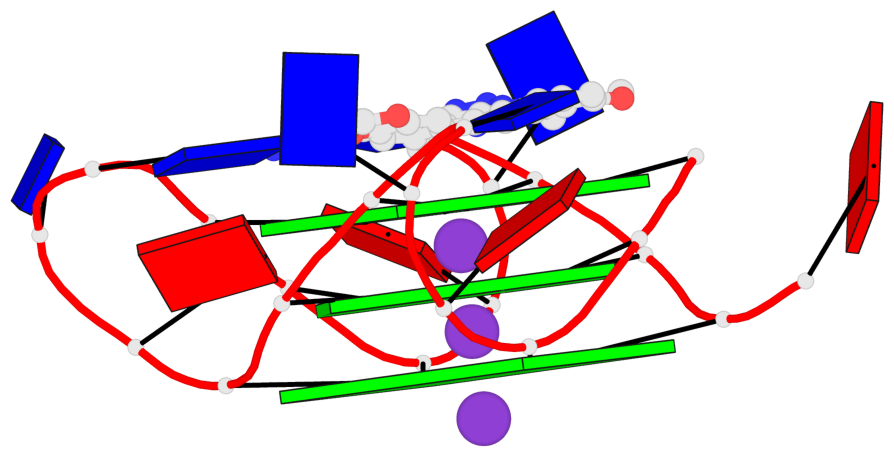
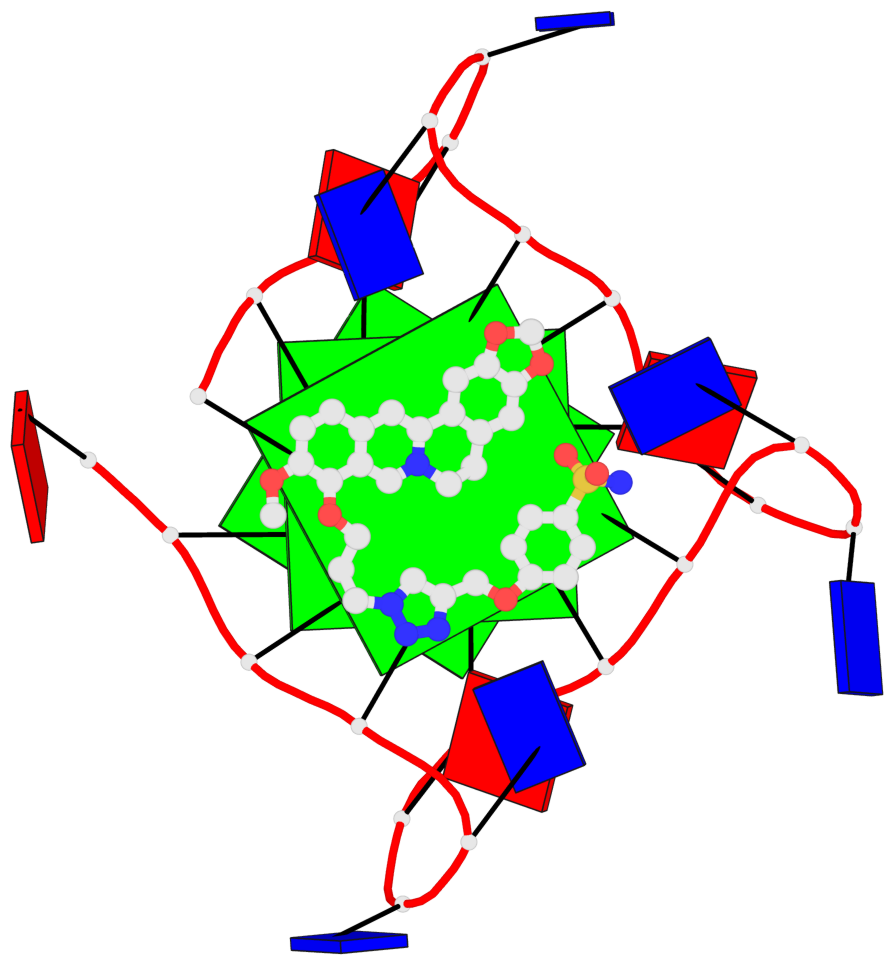
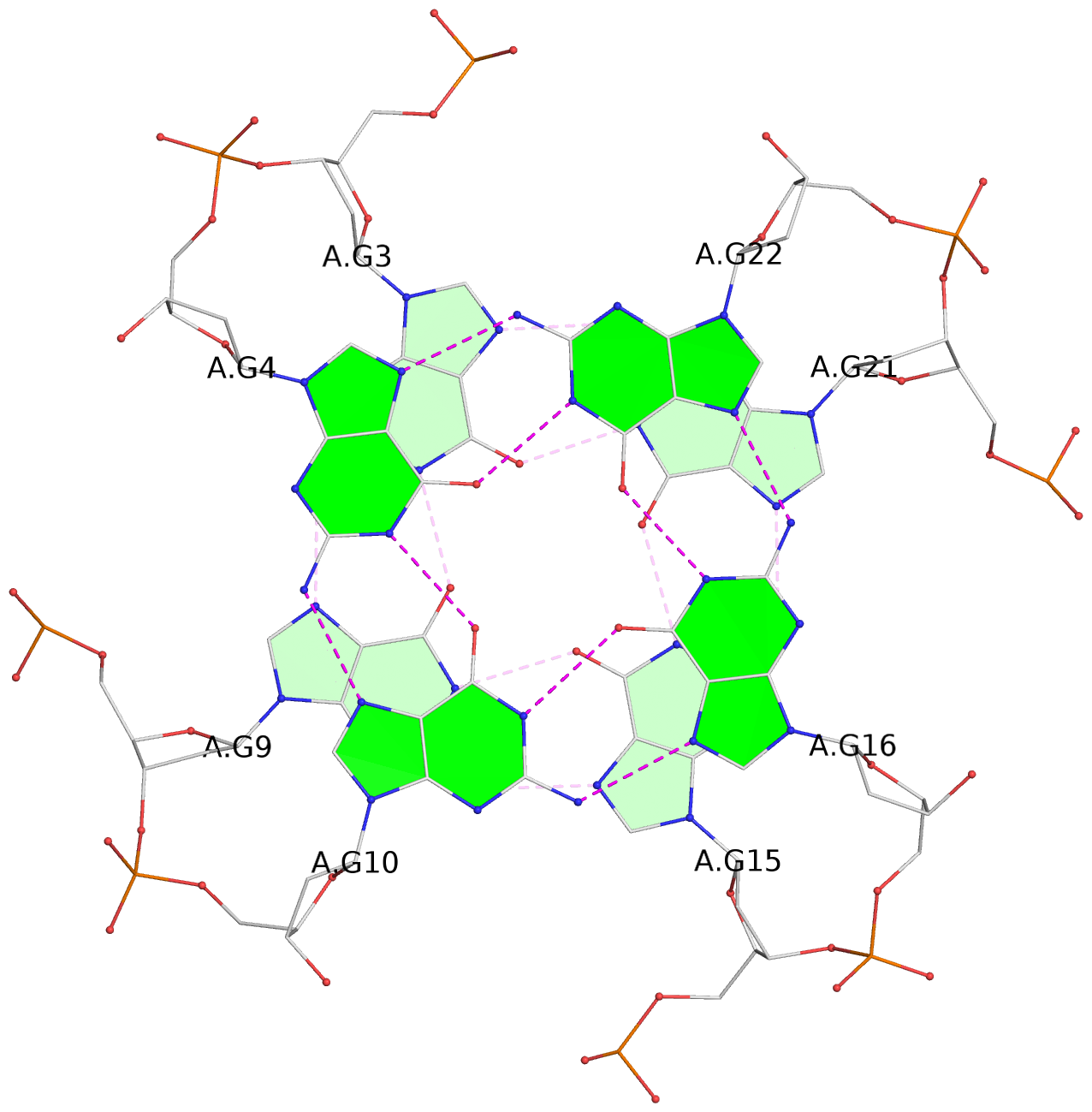
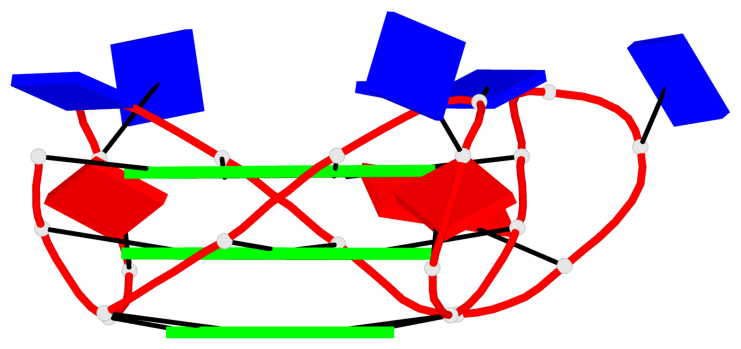
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-P-P-P), parallel(4+0), UUUU
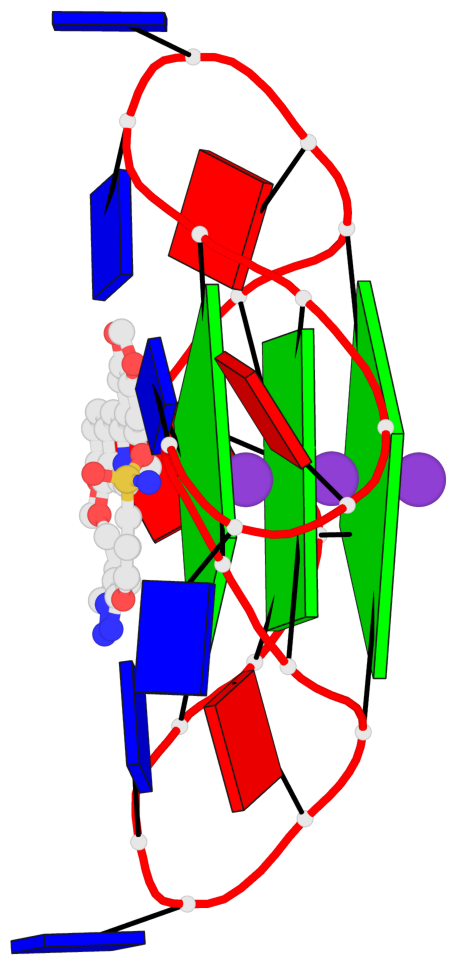
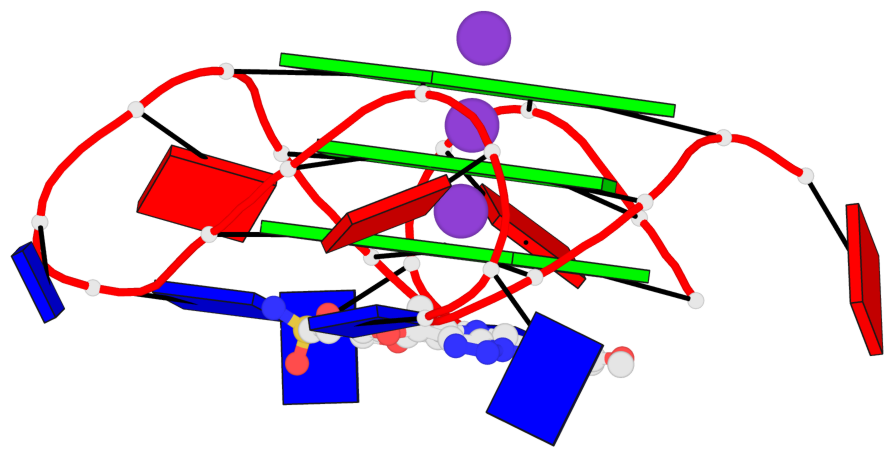
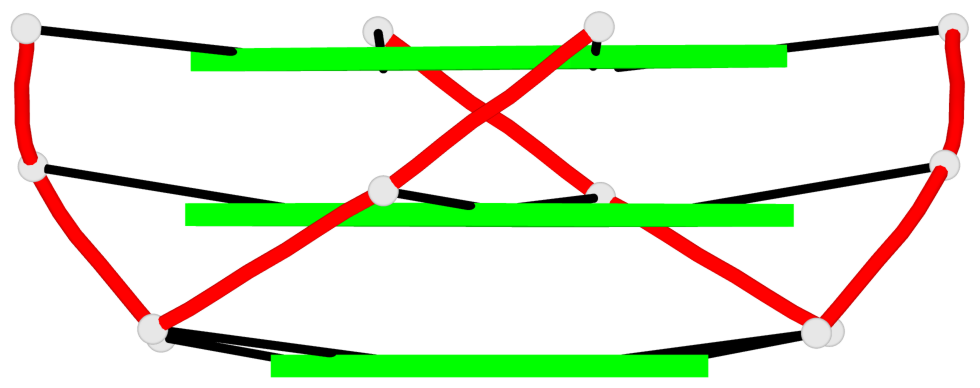
Base-block schematics in six views
List of 3 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.055 type=planar nts=4 GGGG A.DG2,A.DG8,A.DG14,A.DG20 2 glyco-bond=---- sugar=---- groove=---- planarity=0.037 type=planar nts=4 GGGG A.DG3,A.DG9,A.DG15,A.DG21 3 glyco-bond=---- sugar=---- groove=---- planarity=0.153 type=planar nts=4 GGGG A.DG4,A.DG10,A.DG16,A.DG22
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.