Detailed DSSR results for the G-quadruplex: PDB entry 8ht7
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 8ht7
- Class
- DNA binding protein
- Method
- NMR
- Summary
- The n-terminal region of cdc6 specifically recognizes human DNA g-quadruplex
- Reference
- Geng Y, Liu C, Xu N, Shi X, Suen MC, Zhou B, Yan B, Wu C, Li H, Song Y, Chen X, Wang Z, Cai Q, Zhu G (2024): "The N-terminal region of Cdc6 specifically recognizes human DNA G-quadruplex." Int.J.Biol.Macromol., 260, 129487. doi: 10.1016/j.ijbiomac.2024.129487.
- Abstract
- Guanine (G)-rich nucleic acid sequences can form diverse G-quadruplex structures located in functionally significant genome regions, exerting regulatory control over essential biological processes, including DNA replication in vivo. During the initiation of DNA replication, Cdc6 is recruited by the origin recognition complex (ORC) to target specific chromosomal DNA sequences. This study reveals that human Cdc6 interacts with G-quadruplex structure through a distinct region within the N-terminal intrinsically disordered region (IDR), encompassing residues 7-20. The binding region assumes a hook-type conformation, as elucidated by the NMR solution structure in complex with htel21T18. Significantly, mutagenesis and in vivo investigations confirm the highly specific nature of Cdc6's recognition of G-quadruplex. This research enhances our understanding of the fundamental mechanism governing the interaction between G-quadruplex and the N-terminal IDR region of Cdc6, shedding light on the intricate regulation of DNA replication processes.
- G4 notes
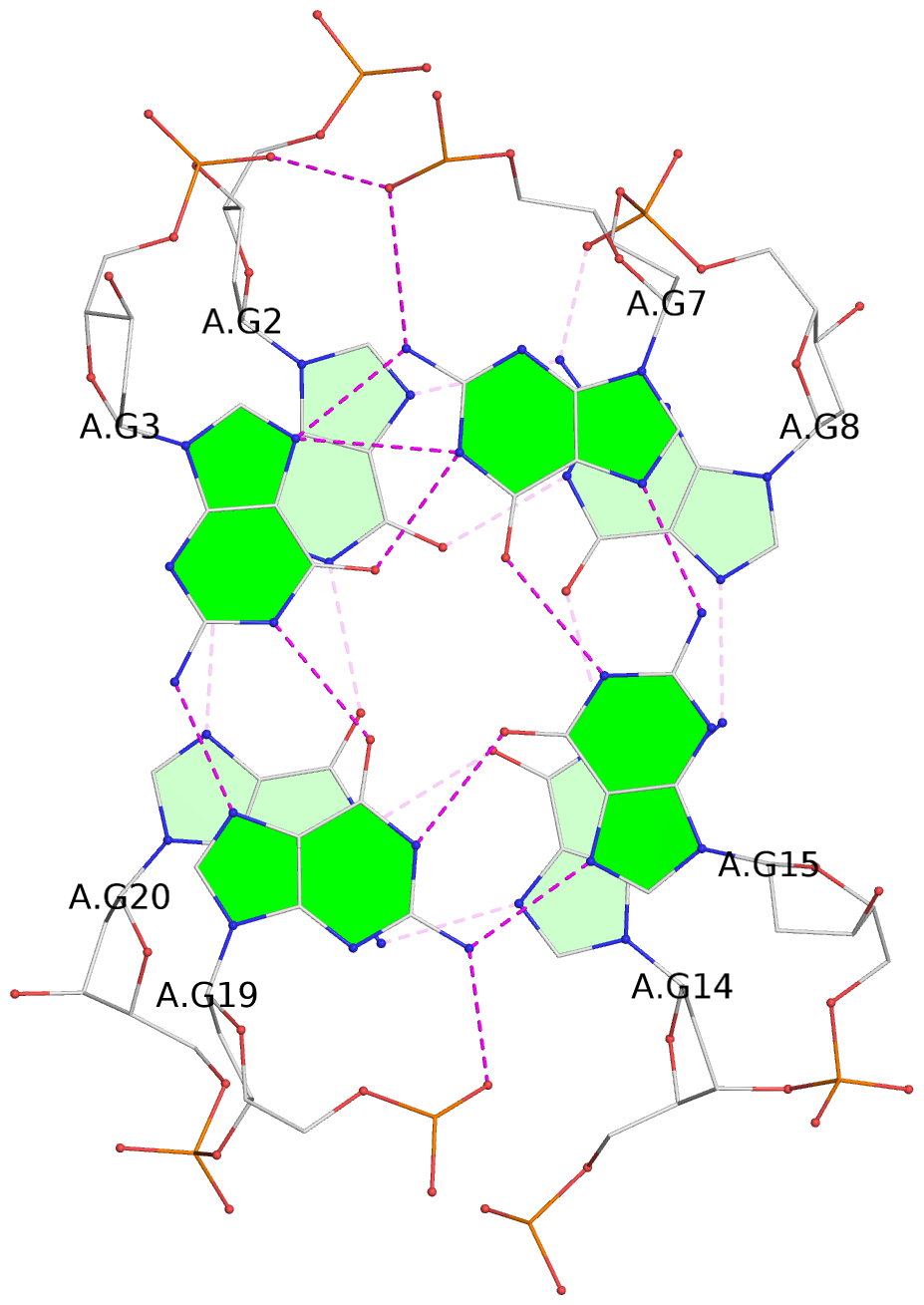
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(+Ln+Lw+Ln), chair(2+2), UDUD
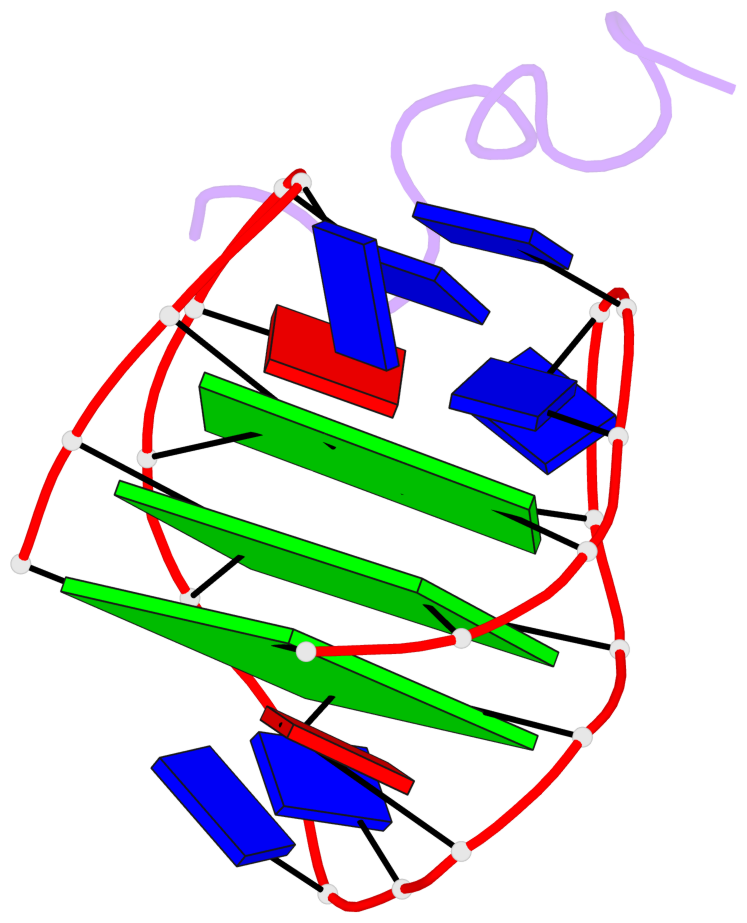
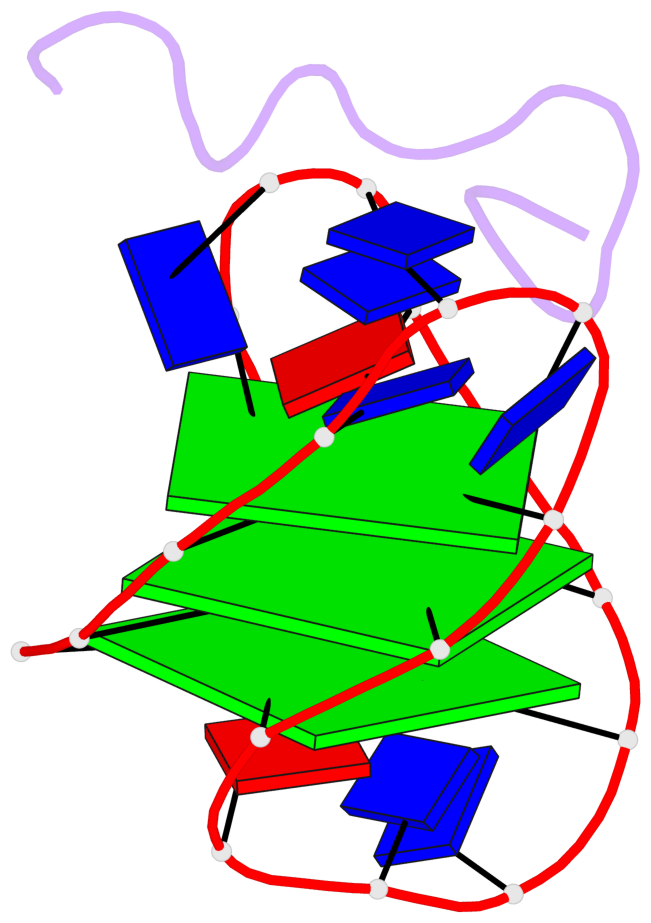
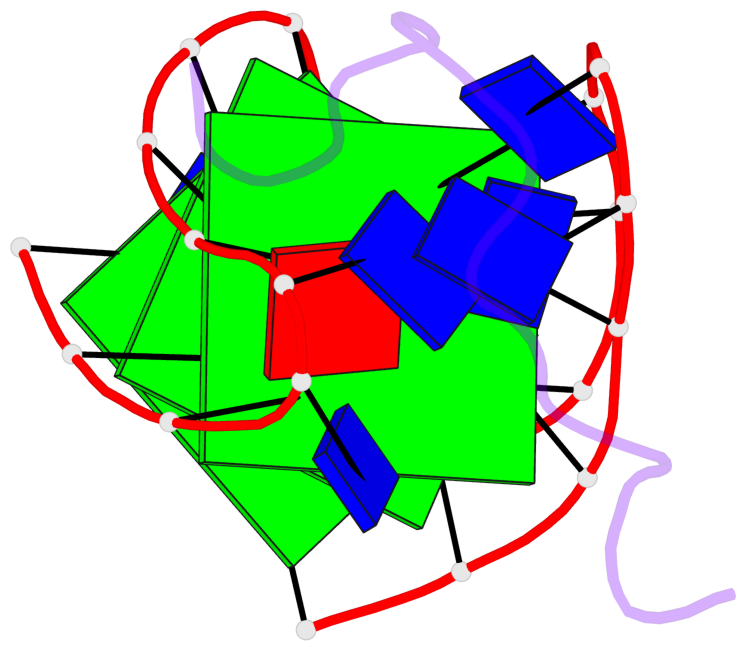
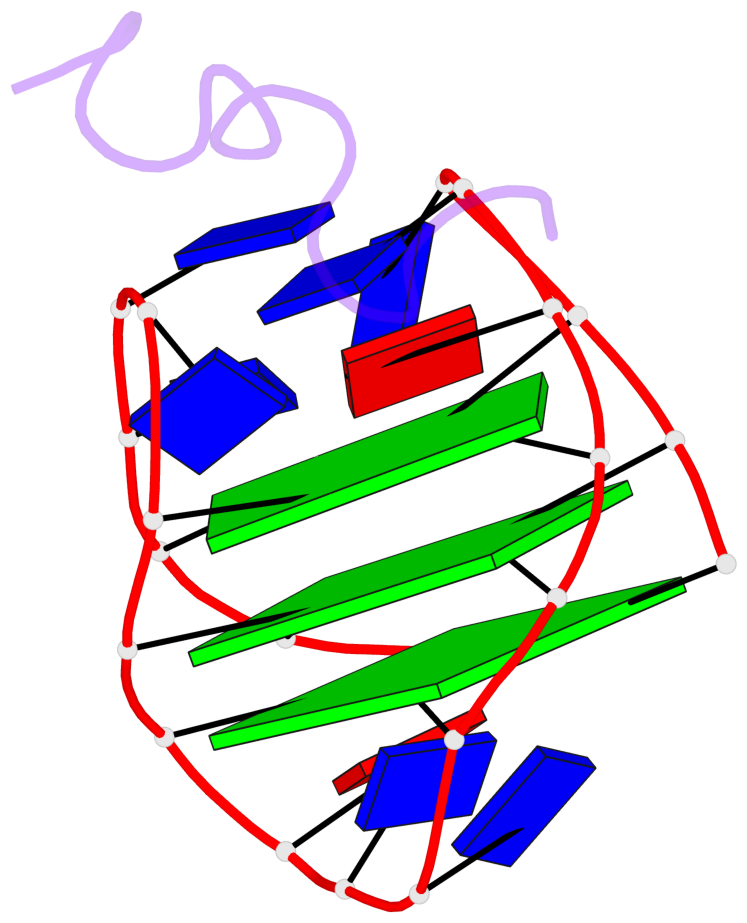
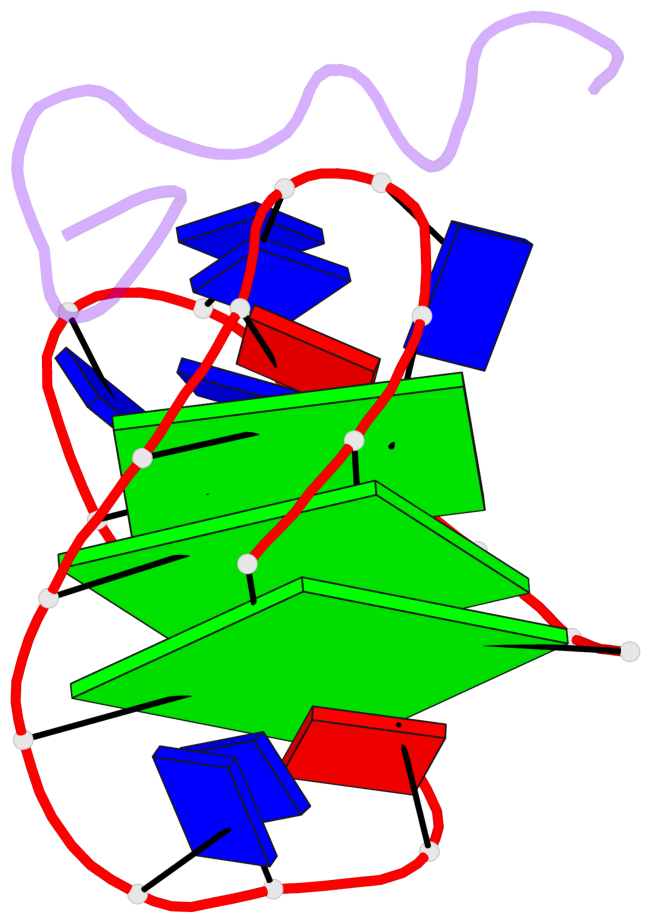
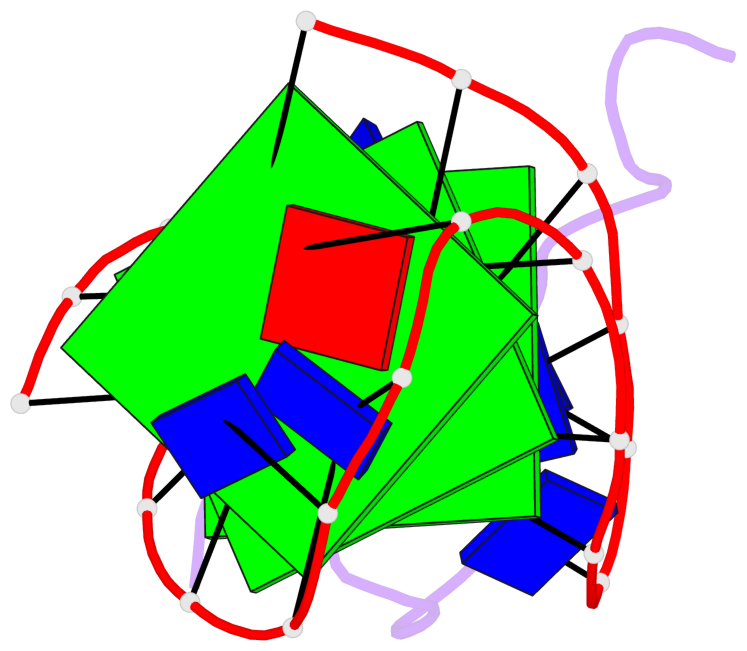
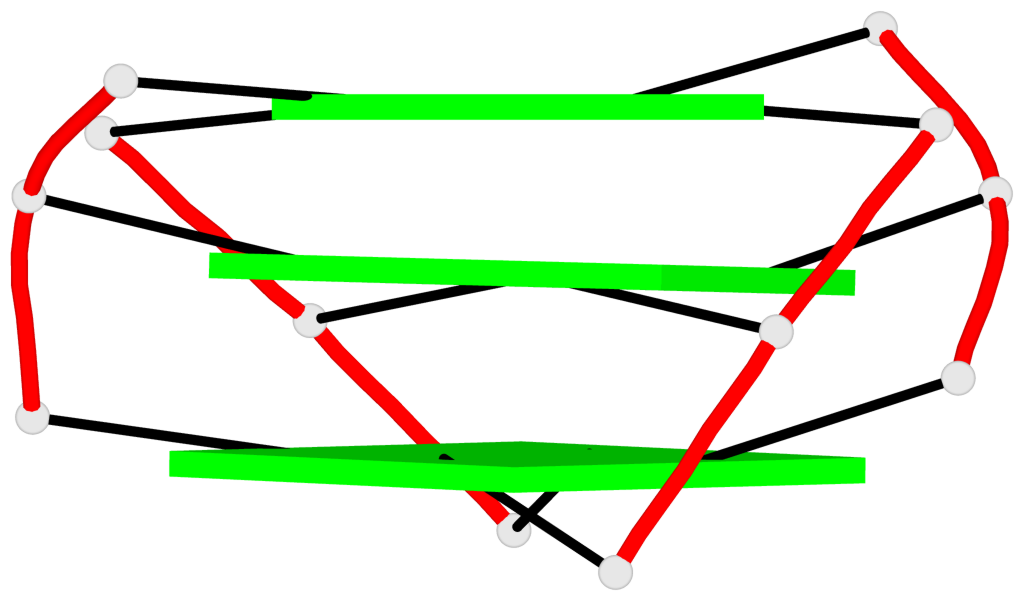
Base-block schematics in six views
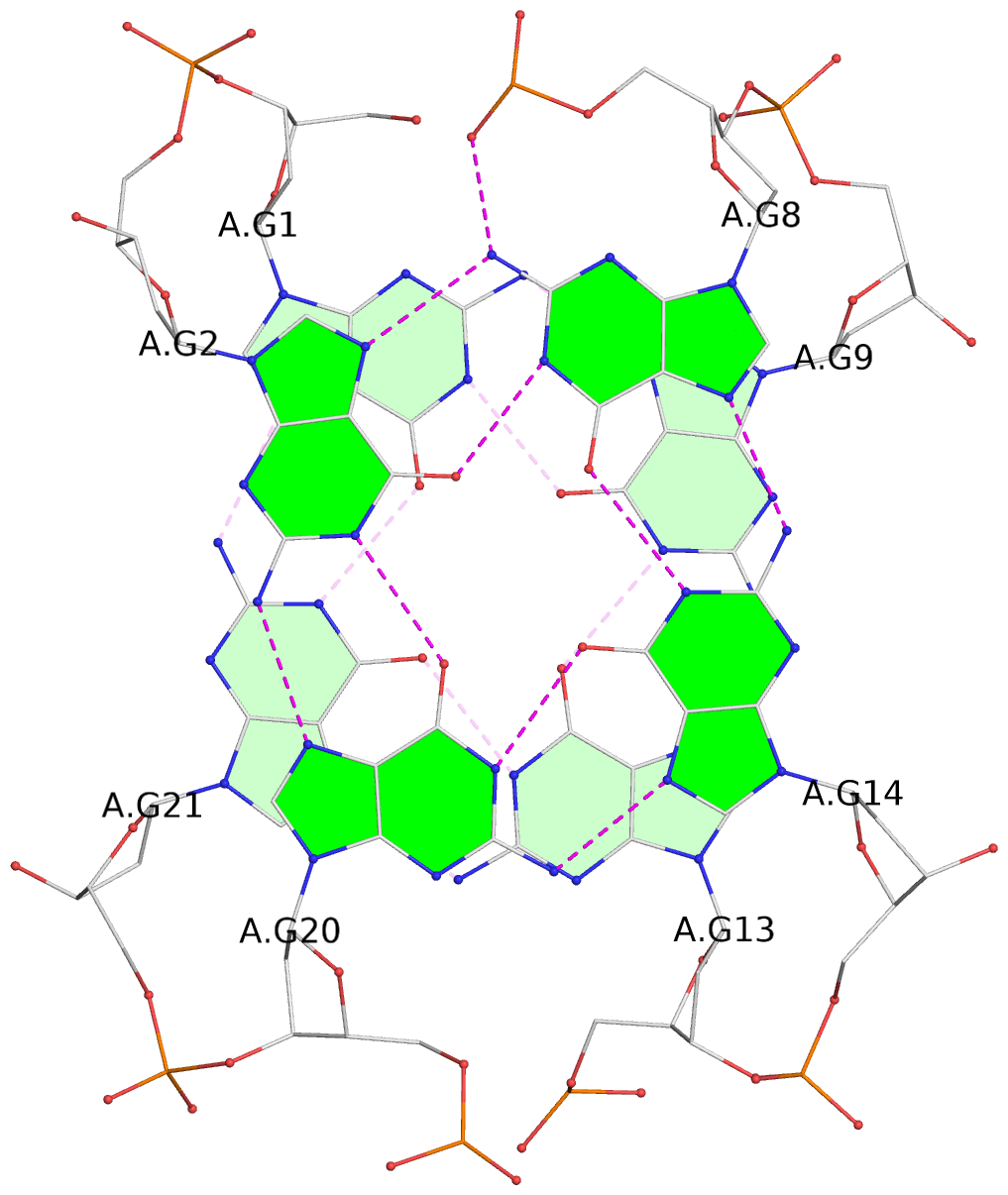
List of 3 G-tetrads
1 glyco-bond=s-s- sugar=---- groove=wnwn planarity=0.077 type=planar nts=4 GGGG A.DG1,A.DG21,A.DG13,A.DG9 2 glyco-bond=-s-s sugar=3--3 groove=wnwn planarity=0.041 type=planar nts=4 GGGG A.DG2,A.DG20,A.DG14,A.DG8 3 glyco-bond=-s-s sugar=3.-3 groove=wnwn planarity=0.218 type=bowl-2 nts=4 GGGG A.DG3,A.DG19,A.DG15,A.DG7
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.