Detailed DSSR results for the G-quadruplex: PDB entry 8kf9
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 8kf9
- Class
- DNA
- Method
- NMR
- Summary
- Stable g-quadruplex conformation formed in promoter region of oncogene ret in 50 mm k+ solution
- Reference
- Zhang YP, Lan WX, Yin SW, Liu ZJ, Xue HJ, Cao CY: "Stable G-quadruplex Conformation Formed in Promoter Region of Oncogene RET Indicates New Target for Anti-cancer Drug Screening."
- Abstract
- G4 notes
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 2(D+PX), UD3(1+3), UDDD
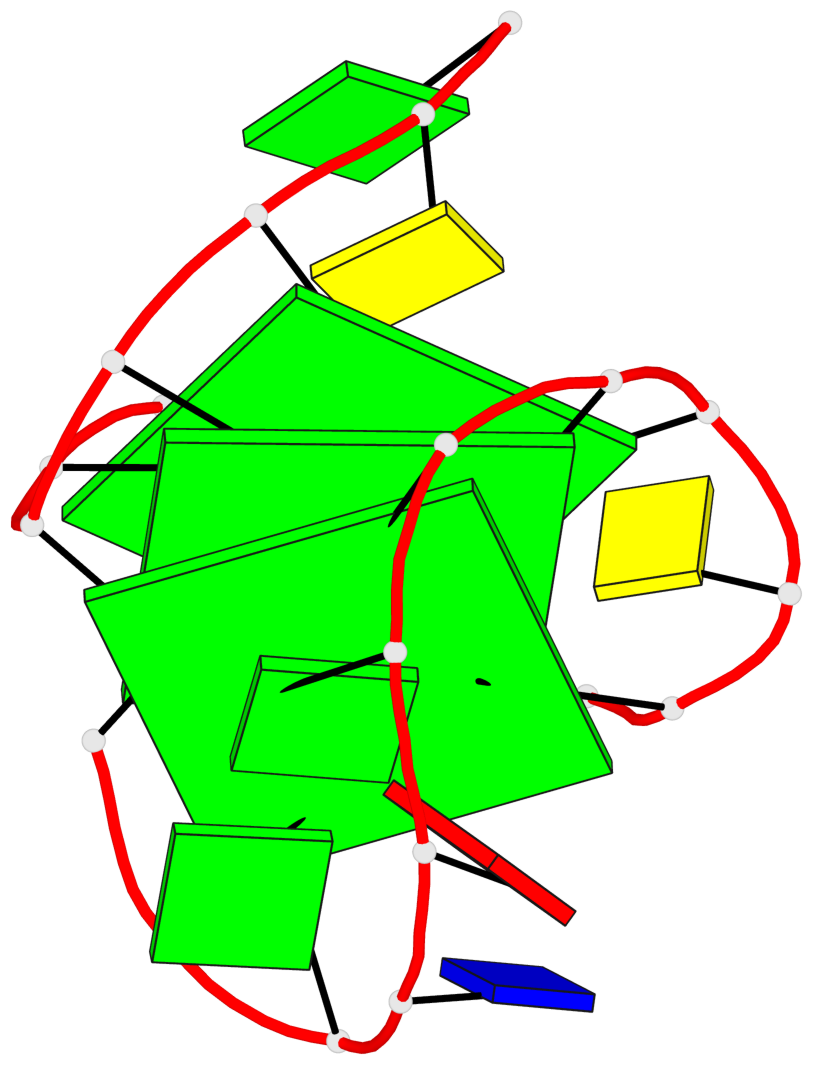
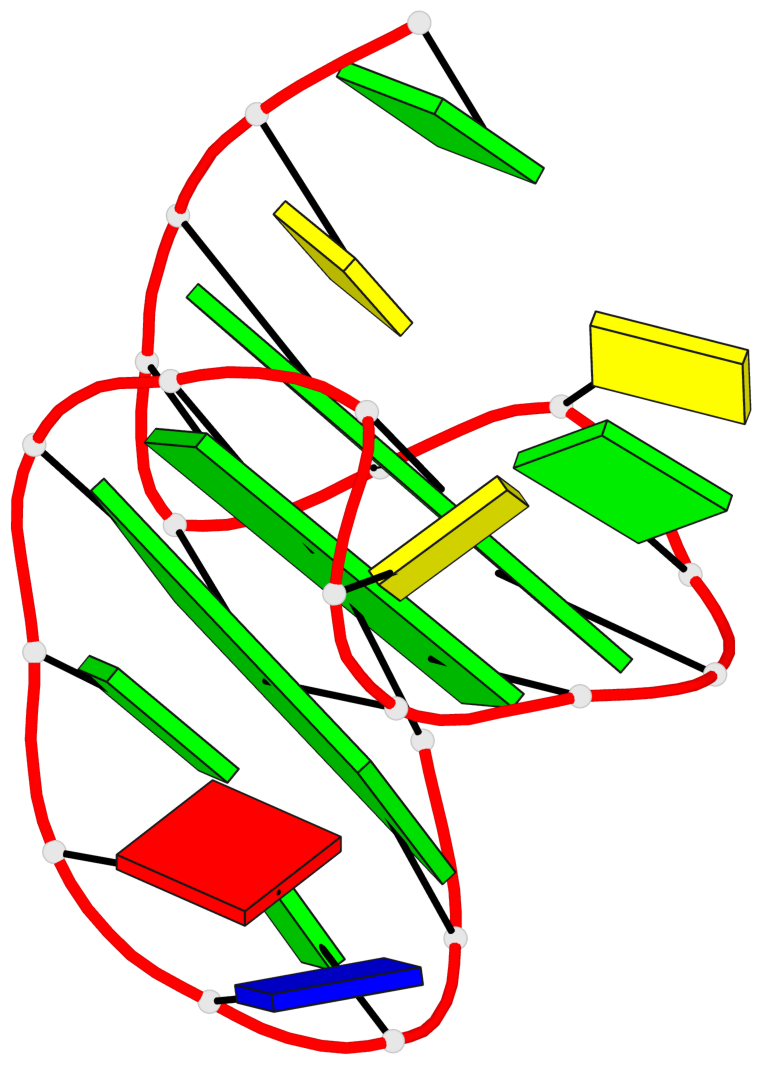
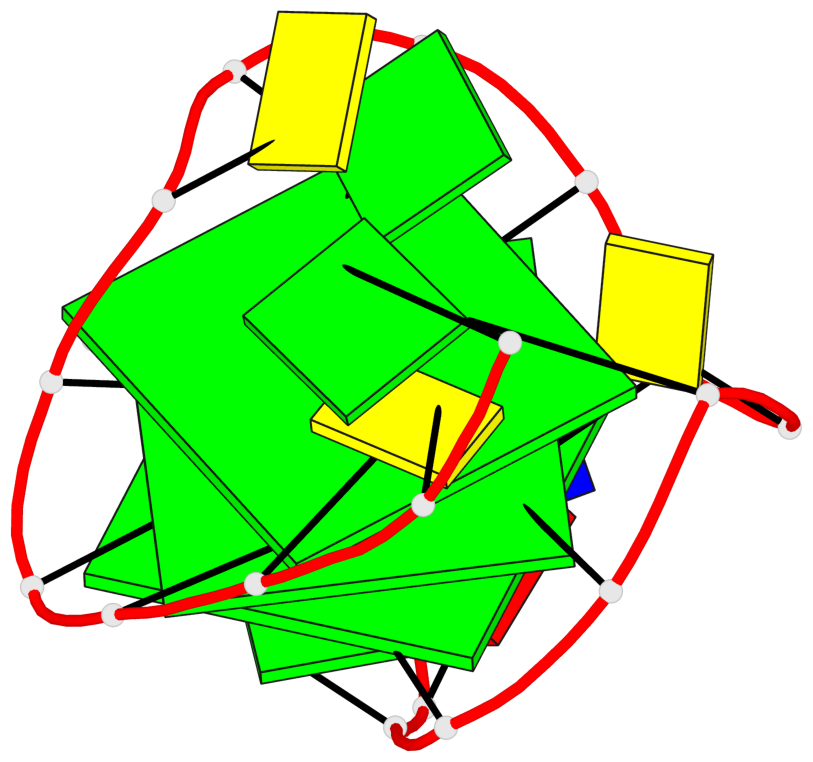
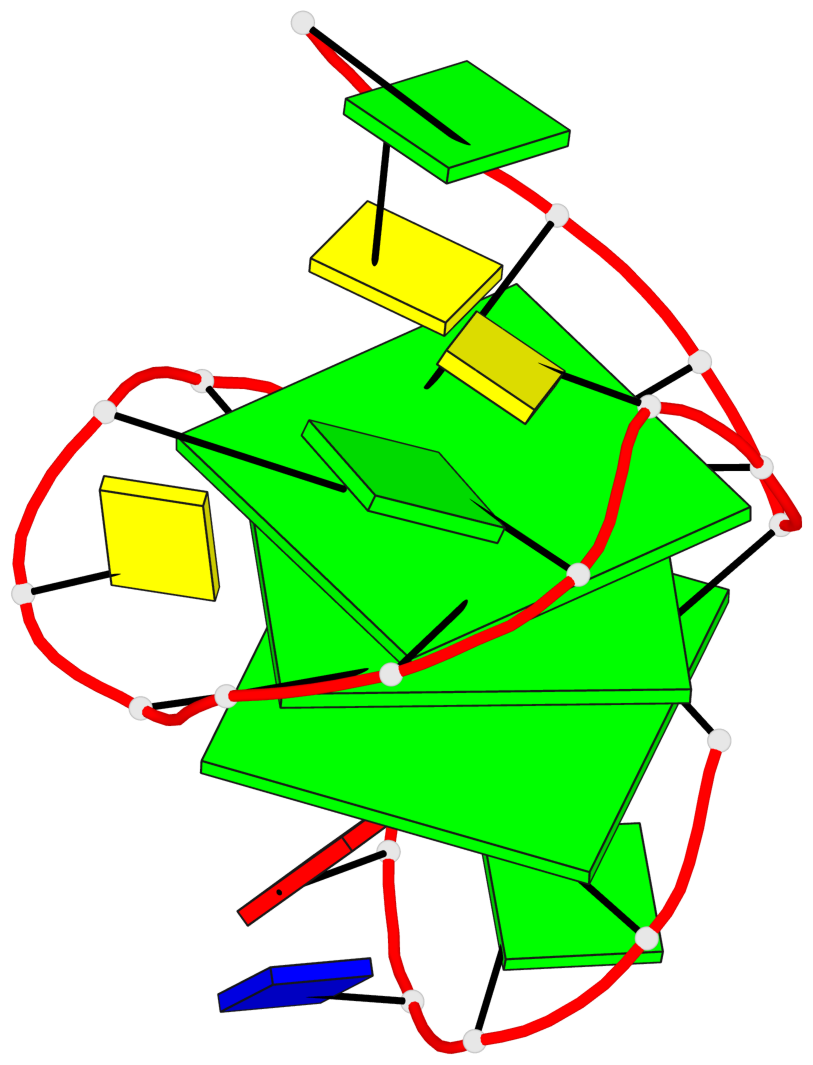
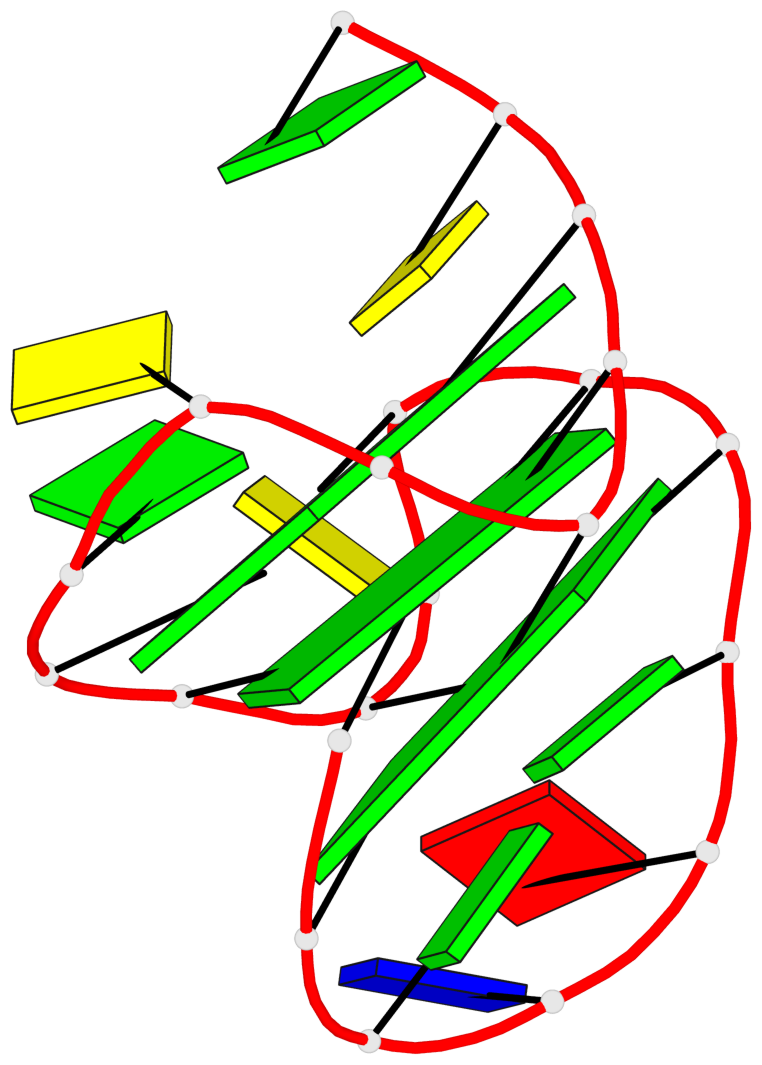
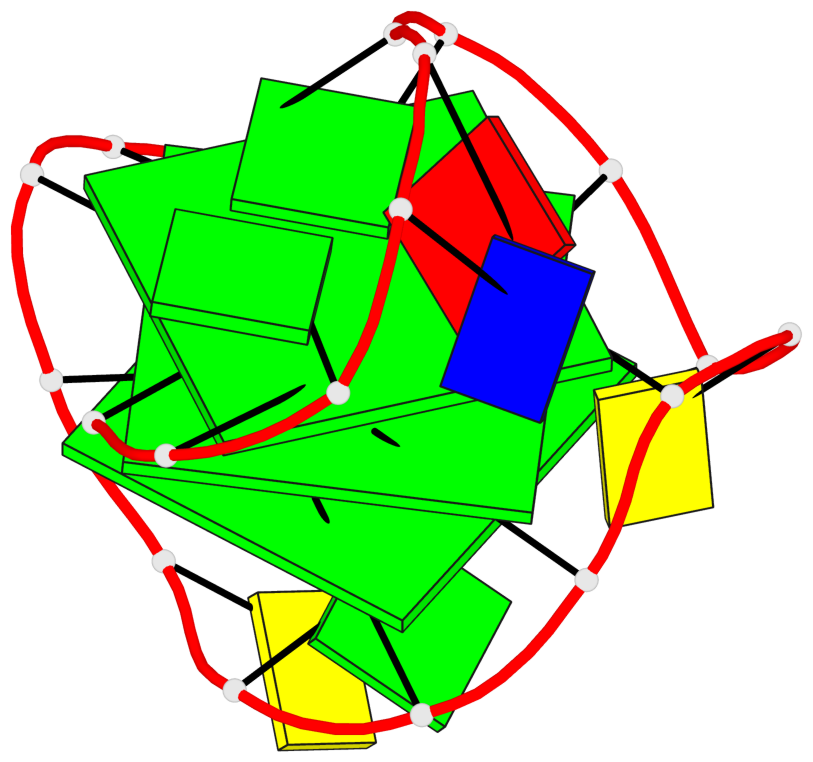
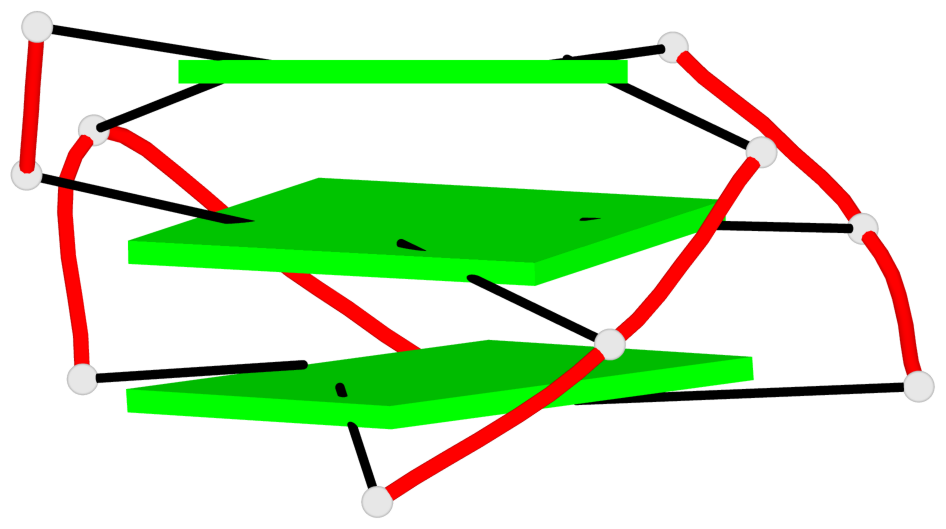
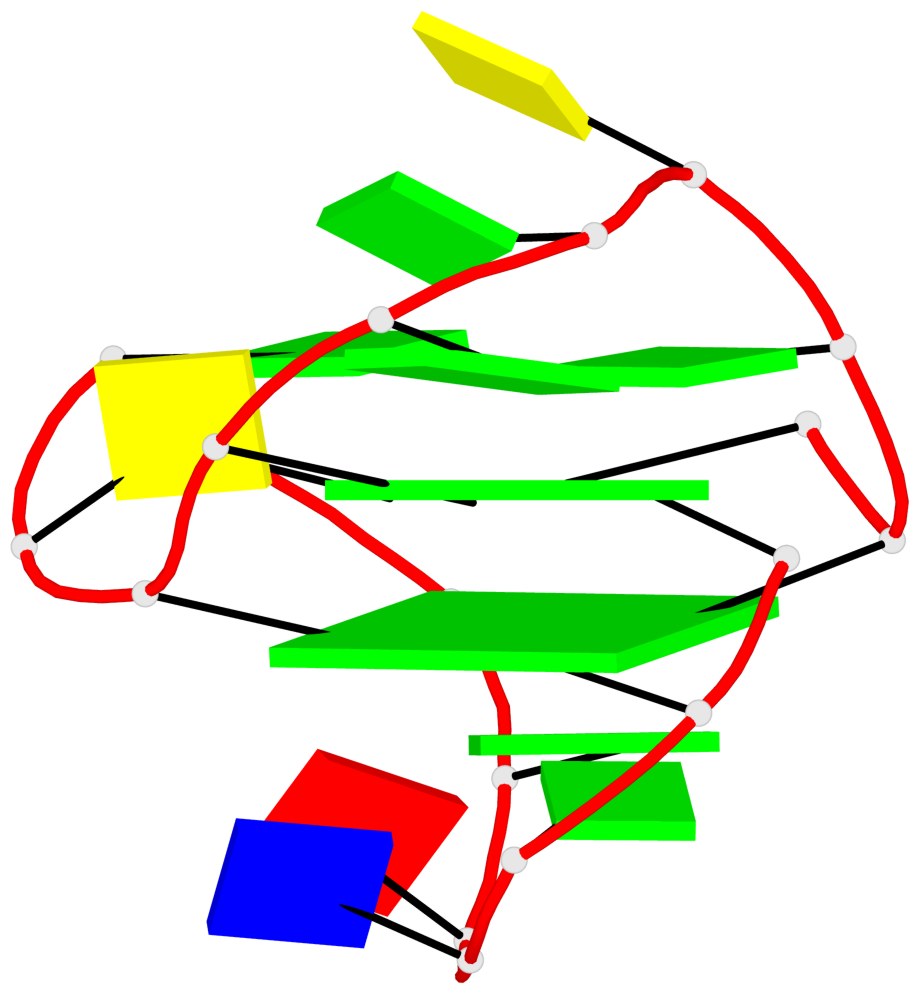
Base-block schematics in six views
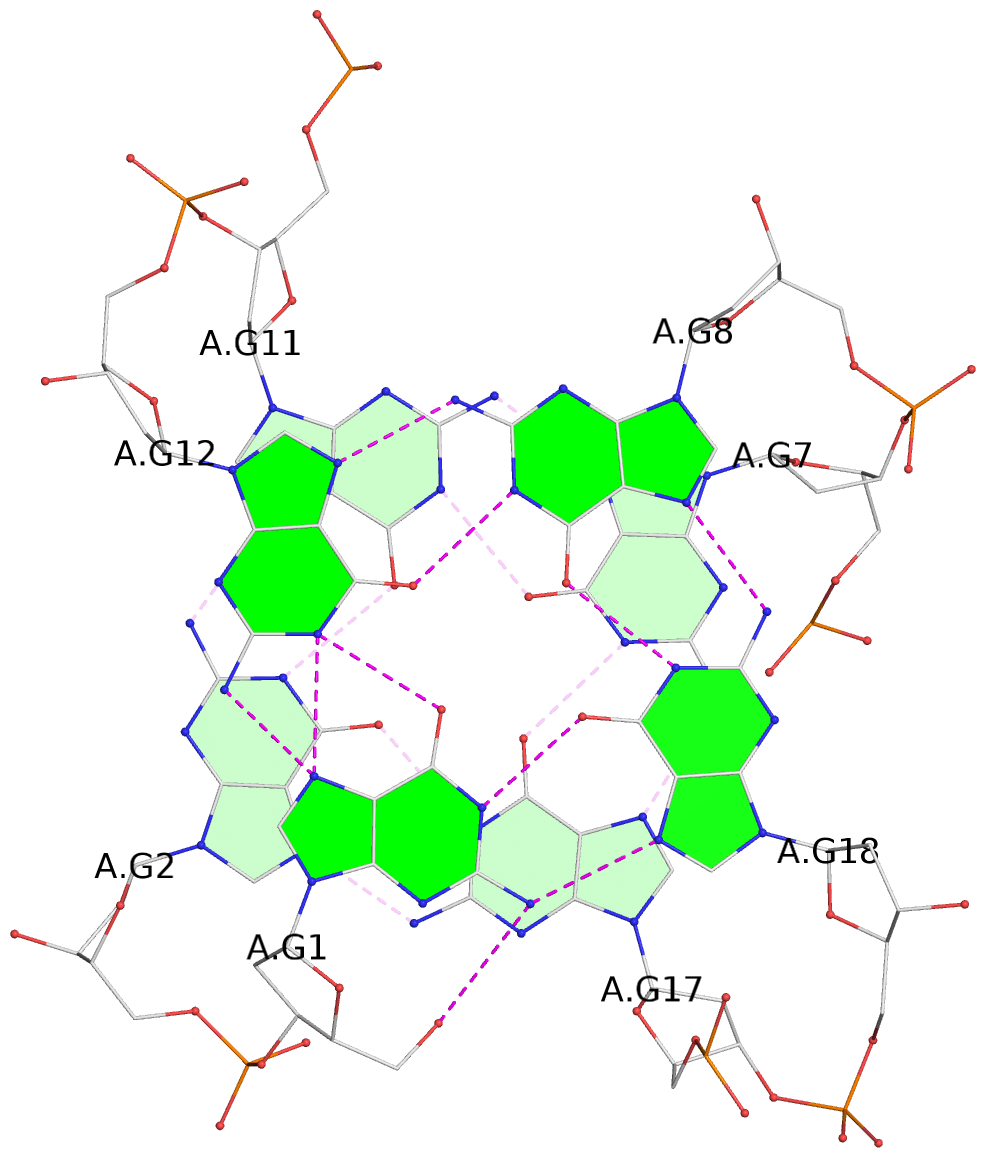
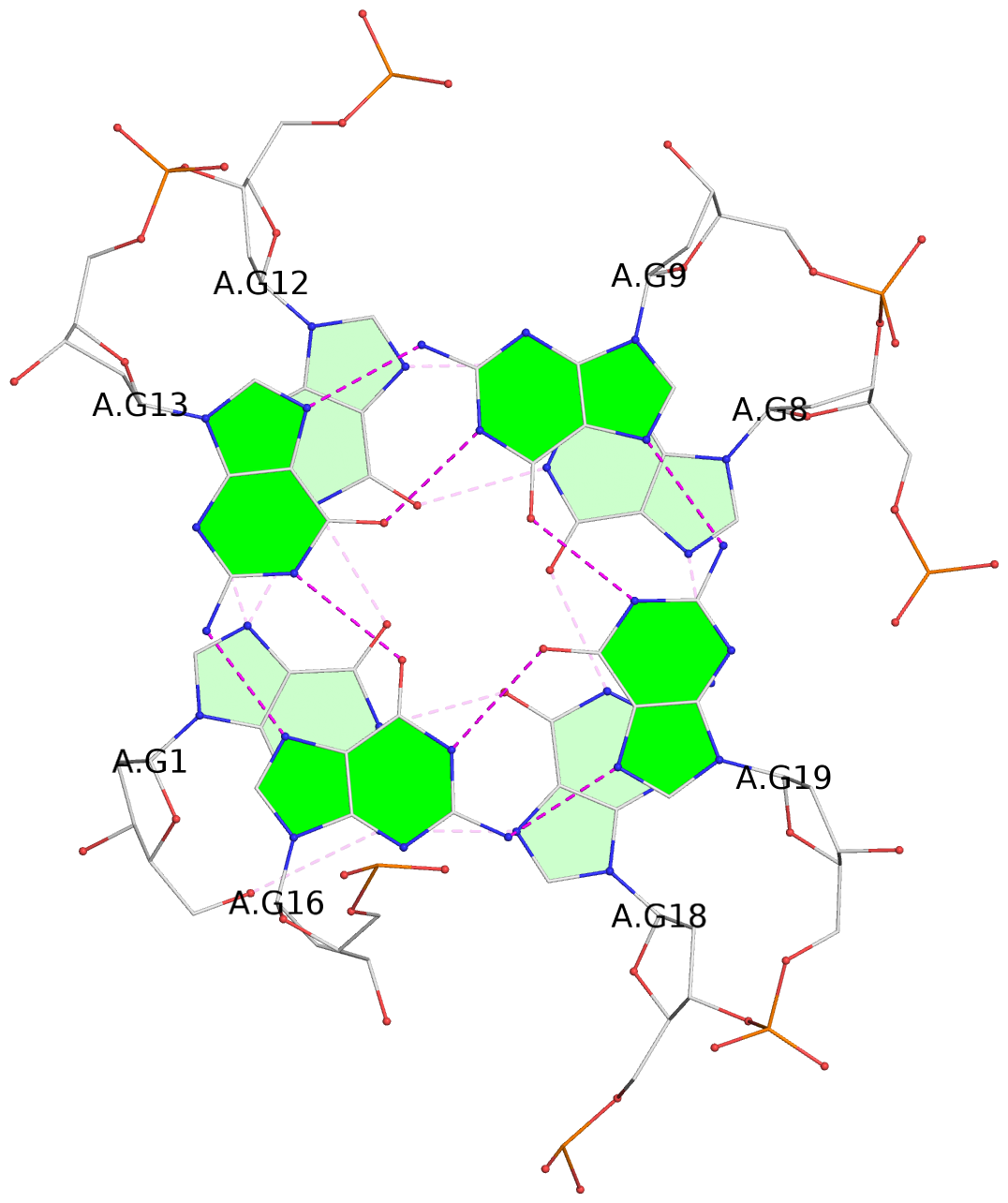
List of 3 G-tetrads
1 glyco-bond=s--- sugar=3.-. groove=w--n planarity=0.193 type=other nts=4 GGGG A.DG1,A.DG12,A.DG8,A.DG18 2 glyco-bond=-ss- sugar=-.-. groove=w-n- planarity=0.196 type=other nts=4 GGGG A.DG2,A.DG11,A.DG7,A.DG17 3 glyco-bond=--s- sugar=--3. groove=-wn- planarity=0.221 type=other nts=4 GGGG A.DG9,A.DG13,A.DG16,A.DG19
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 2 G-tetrad layers, 3 loops, INTRA-molecular, UDDD, hybrid-(mixed), 2(D+PX), UD3(1+3)
List of 2 non-stem G4-loops (including the two closing Gs)
1 type=lateral helix=#1 nts=4 GGCG A.DG13,A.DG14,A.DC15,A.DG16 2 type=V-shaped helix=#1 nts=4 GGGG A.DG16,A.DG17,A.DG18,A.DG19