Detailed DSSR results for the G-quadruplex: PDB entry 8rw2
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 8rw2
- Class
- DNA
- Method
- NMR
- Summary
- Structure of a chair-type antiparallel quadruplex-duplex hybrid at ph 6
- Reference
- Vianney YM, Jana J, Weisz K (2024): "A pH-Responsive Topological Switch Based on a DNA Quadruplex-Duplex Hybrid." Chemistry, 30, e202400722. doi: 10.1002/chem.202400722.
- Abstract
- A guanine-rich oligonucleotide based on a human telomeric sequence but with the first three-nucleotide intervening stretch replaced by a putative 15-nucleotide hairpin-forming sequence shows a pH-dependent folding into different quadruplex-duplex hybrids in a potassium containing buffer. At slightly acidic pH, the quadruplex domain adopts a chair-type conformation. Upon increasing the pH, a transition with a midpoint close to neutral pH to a major and minor (3+1) hybrid topology with either a coaxially stacked or orthogonally oriented duplex stem-loop occurs. NMR-derived high-resolution structures reveal that an adenine protonation is prerequisite for the formation of a non-canonical base quartet, capping the outer G-tetrad at the quadruplex-duplex interface and stabilizing the antiparallel chair conformation in an acidic environment. Being directly associated with interactions at the quadruplex-duplex interface, this unique pH-dependent topological transition is fully reversible. Coupled with a conformation-sensitive optical readout demonstrated as a proof of concept using the fluorescent dye thiazole orange, the present quadruplex-duplex hybrid architecture represents a potentially valuable pH-sensing system responsive in a physiological pH range of 7±1.
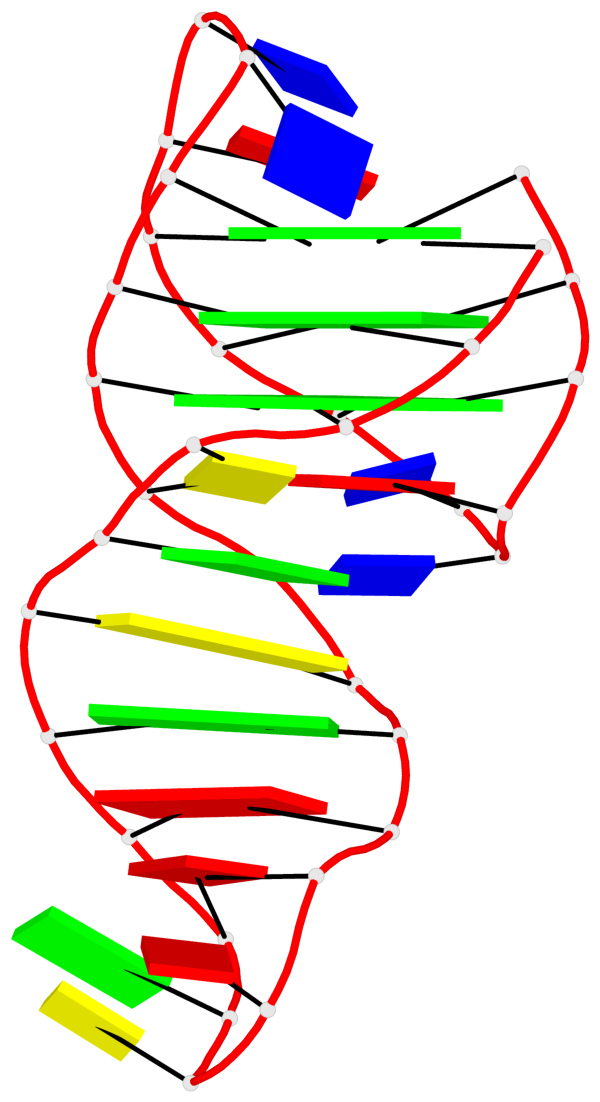
- G4 notes
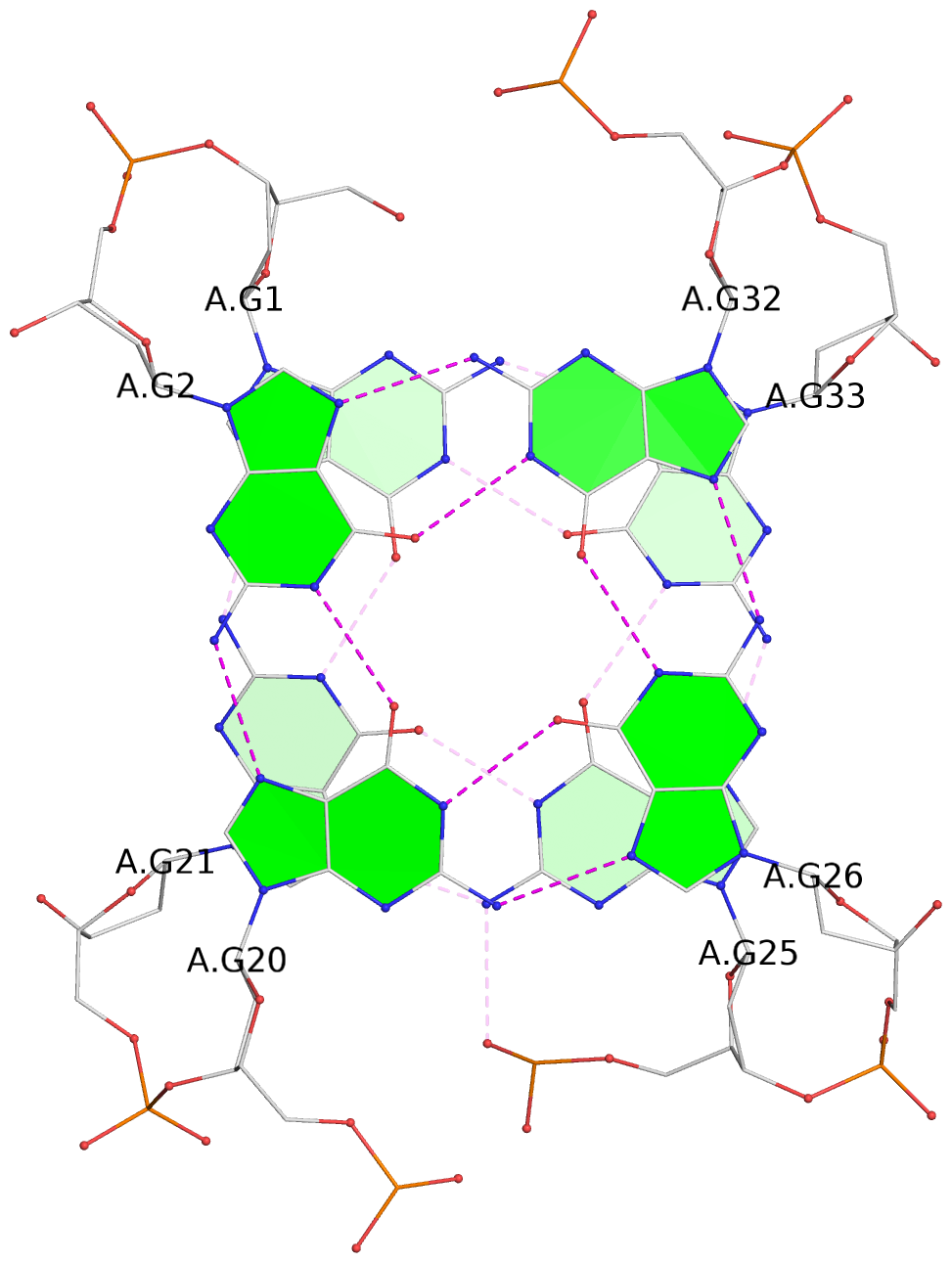
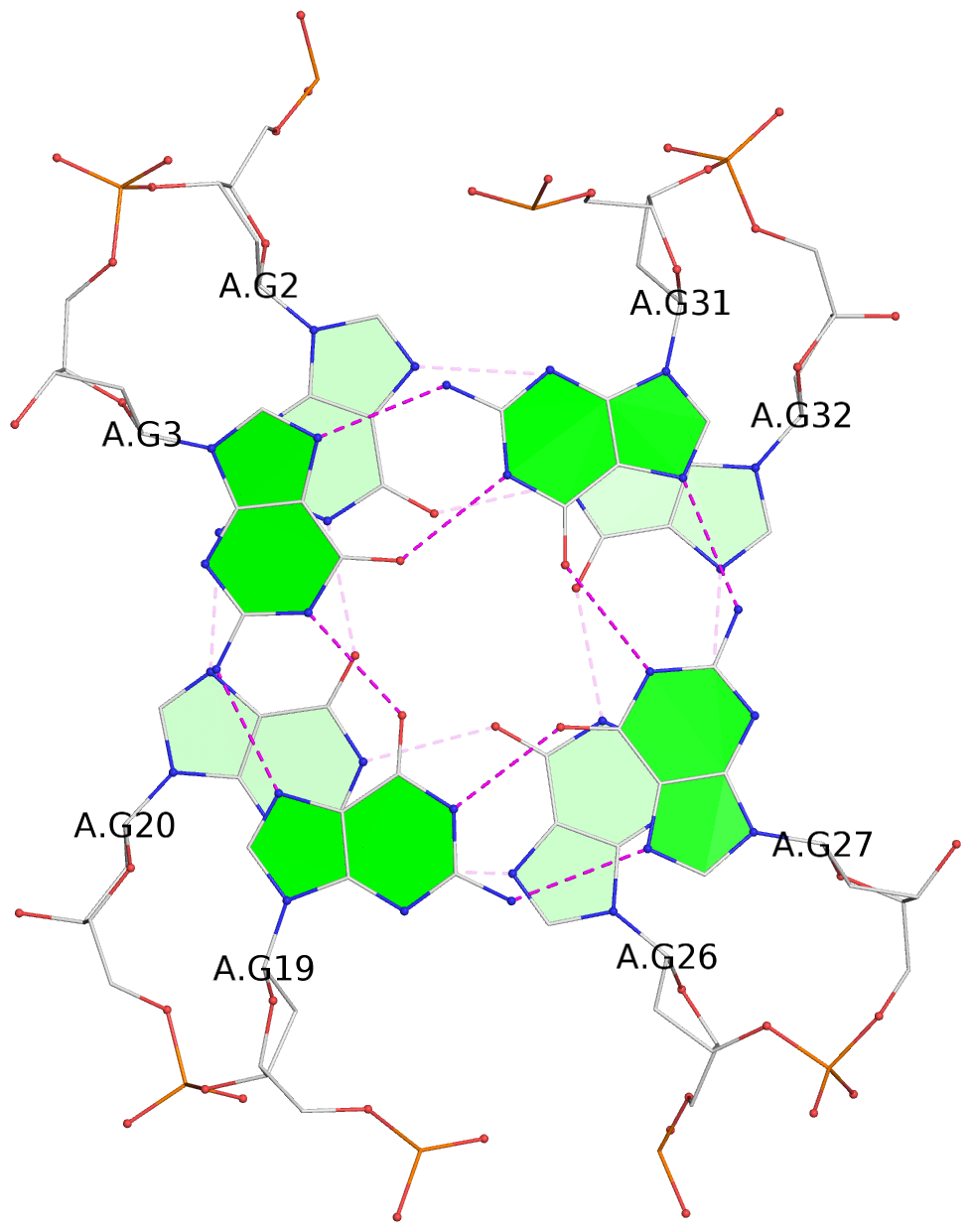
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-Lw-Ln-Lw), chair(2+2), UDUD
List of 3 G-tetrads
1 glyco-bond=s-s- sugar=---- groove=wnwn planarity=0.530 type=bowl nts=4 GGGG A.DG1,A.DG21,A.DG25,A.DG33 2 glyco-bond=-s-s sugar=---- groove=wnwn planarity=0.346 type=other nts=4 GGGG A.DG2,A.DG20,A.DG26,A.DG32 3 glyco-bond=-s-s sugar=---- groove=wnwn planarity=0.289 type=saddle nts=4 GGGG A.DG3,A.DG19,A.DG27,A.DG31
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.