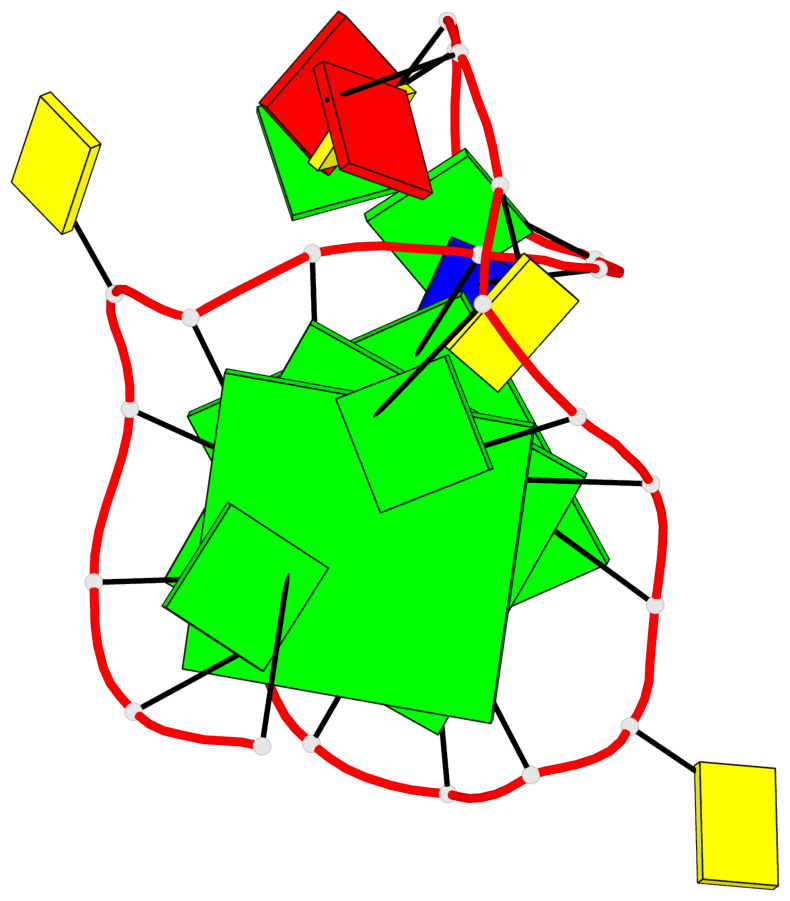
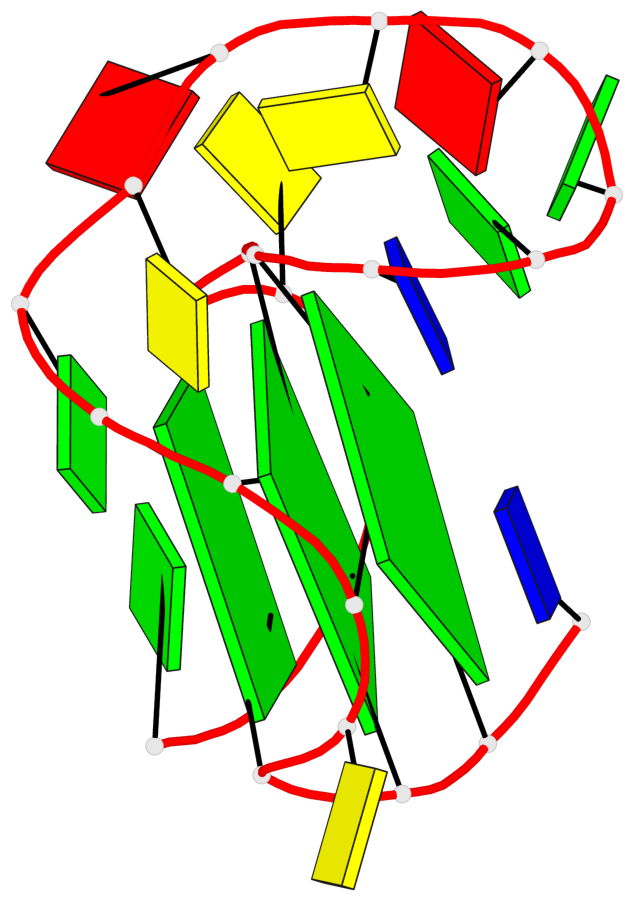
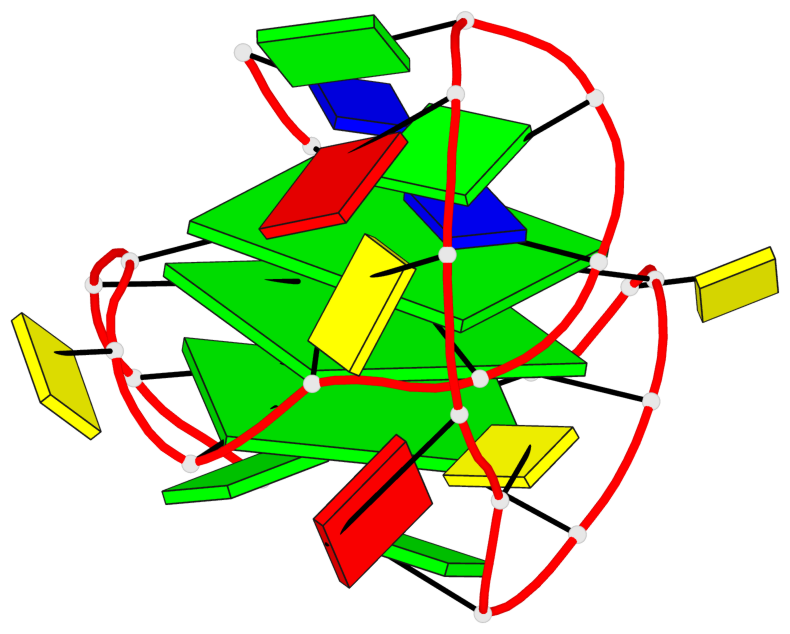
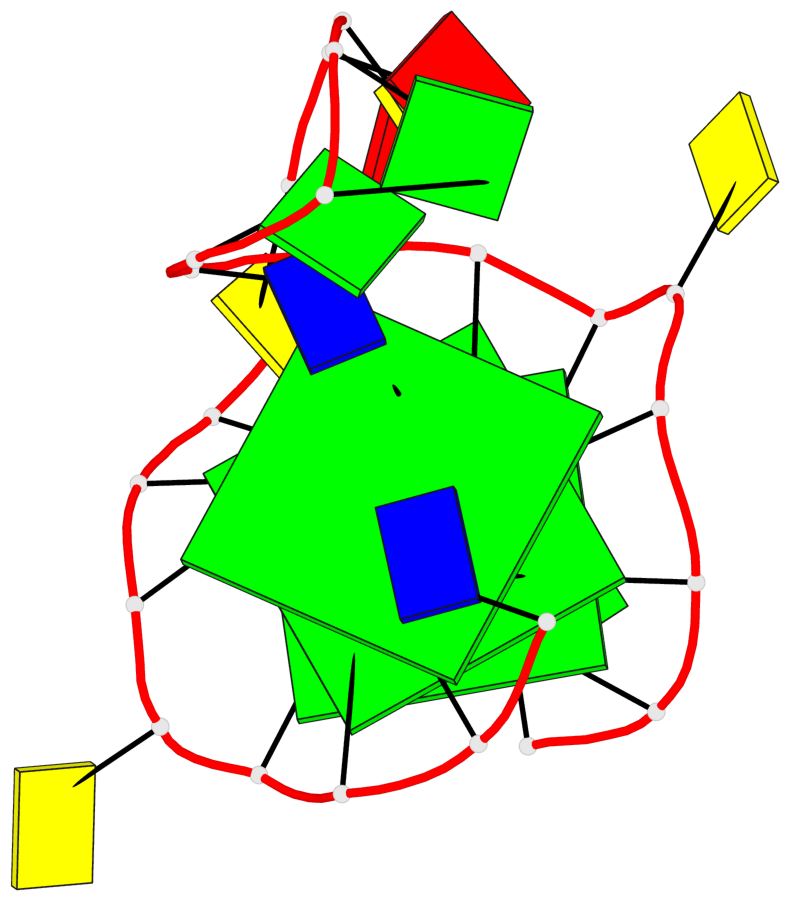
Detailed DSSR results for the G-quadruplex: PDB entry 8rzx
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 8rzx
- Class
- DNA
- Method
- NMR
- Summary
- Solution structure of a parallel stranded g-quadruplex formed in orai1 promoter
- Reference
- Chatterjee O, Jana J, Panda S, Dutta A, Sharma A, Saurav S, Motiani RK, Weisz K, Chatterjee S (2024): "Remodeling Ca 2+ dynamics by targeting a promising E-box containing G-quadruplex at ORAI1 promoter in triple-negative breast cancer." Cell Calcium, 123, 102944. doi: 10.1016/j.ceca.2024.102944.
- Abstract
- ORAI1 is an intrinsic component of store-operated calcium entry (SOCE) that strictly regulates Ca2+ influx in most non-excitable cells. ORAI1 is overexpressed in a wide variety of cancers, and its signal transduction has been associated with chemotherapy resistance. There is extensive proteomic interaction of ORAI1 with other channels and effectors, resulting in various altered phenotypes. However, the transcription regulation of ORAI1 is not well understood. We have found a putative G-quadruplex (G4) motif, ORAI1-Pu, in the upstream promoter region of the gene, having regulatory functions. High-resolution 3-D NMR structure elucidation suggests that ORAI1-Pu is a stable parallel-stranded G4, having a long 8-nt loop imparting dynamics without affecting the structural stability. The protruded loop further houses an E-box motif that provides a docking site for transcription factors like Zeb1. The G4 structure was also endogenously observed using Chromatin Immunoprecipitation (ChIP) with anti-G4 antibody (BG4) in the MDA-MB-231 cell line overexpressing ORAI1. Ligand-mediated stabilization suggested that the stabilized G4 represses transcription in cancer cell line MDA-MB-231. Downregulation of transcription further led to decreased Ca2+ entry by the SOCE pathway, as observed by live-cell Fura-2 Ca2+ imaging.
- G4 notes
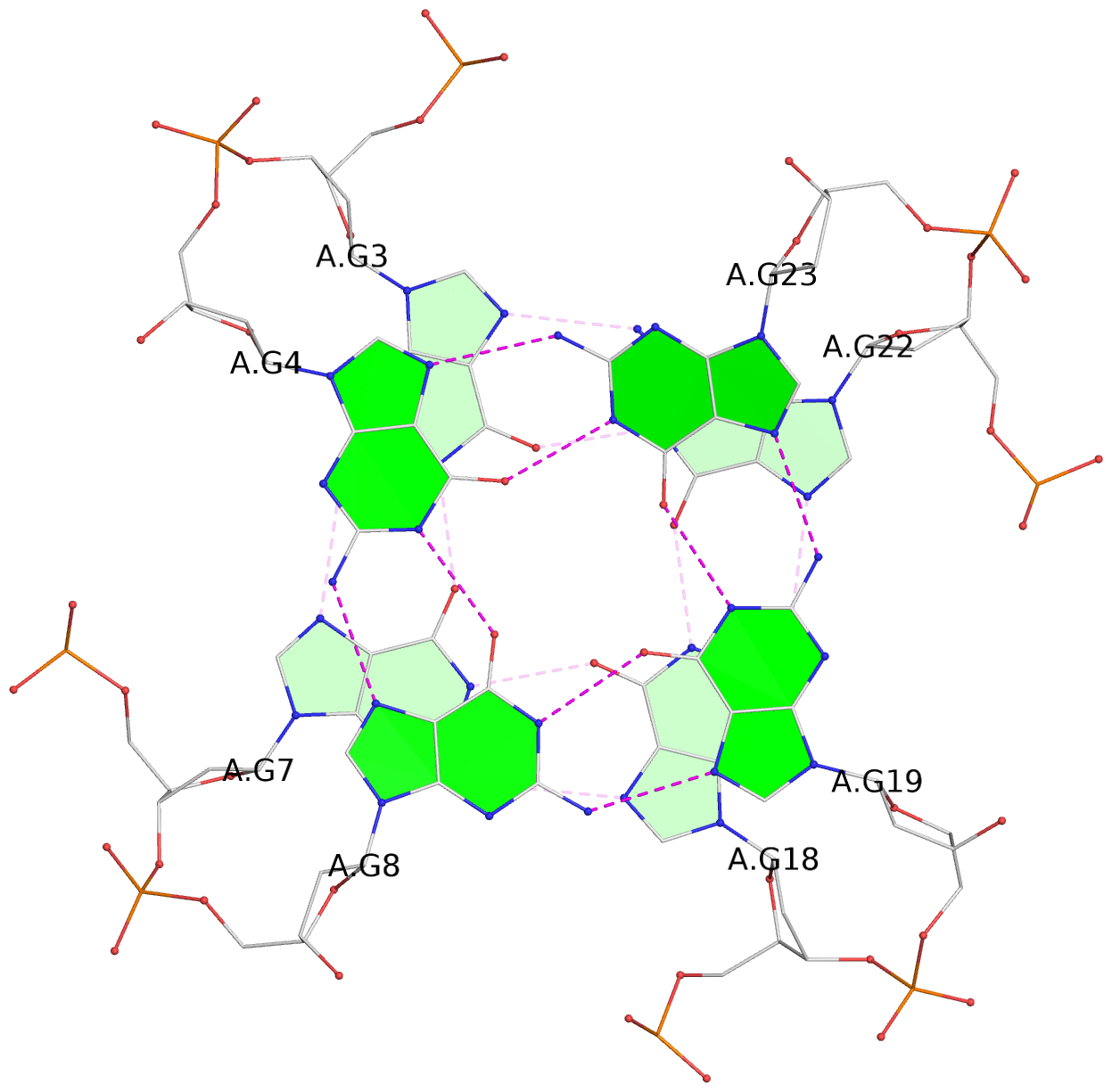
- 3 G-tetrads, 1 G4 helix, 1 G4 stem, 3(-P-P-P), parallel(4+0), UUUU
Base-block schematics in six views
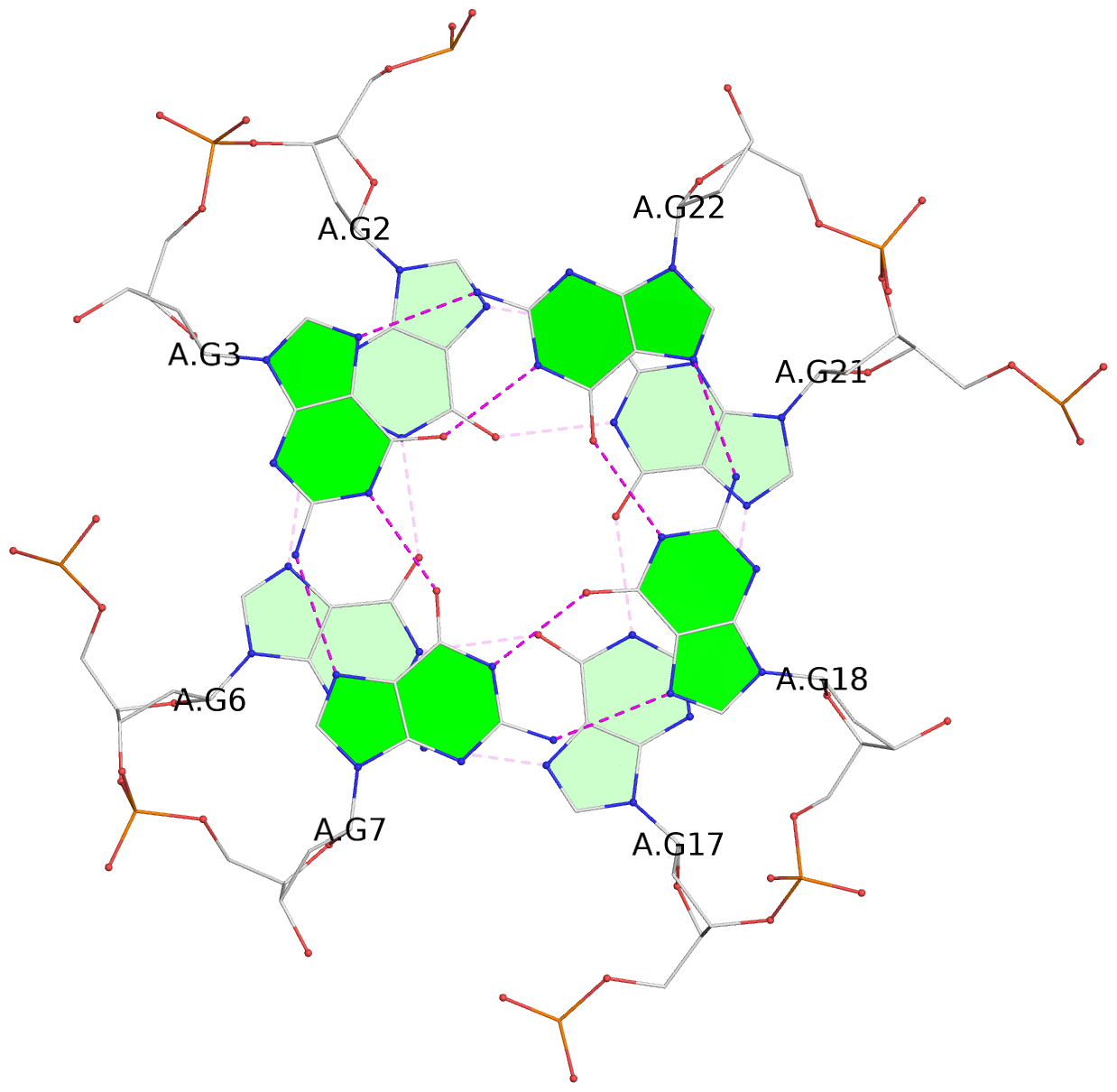
List of 3 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.128 type=planar nts=4 GGGG A.DG2,A.DG6,A.DG17,A.DG21 2 glyco-bond=---- sugar=---- groove=---- planarity=0.218 type=other nts=4 GGGG A.DG3,A.DG7,A.DG18,A.DG22 3 glyco-bond=---- sugar=---- groove=---- planarity=0.336 type=other nts=4 GGGG A.DG4,A.DG8,A.DG19,A.DG23
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrad layers, INTRA-molecular, with 1 stem
List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.