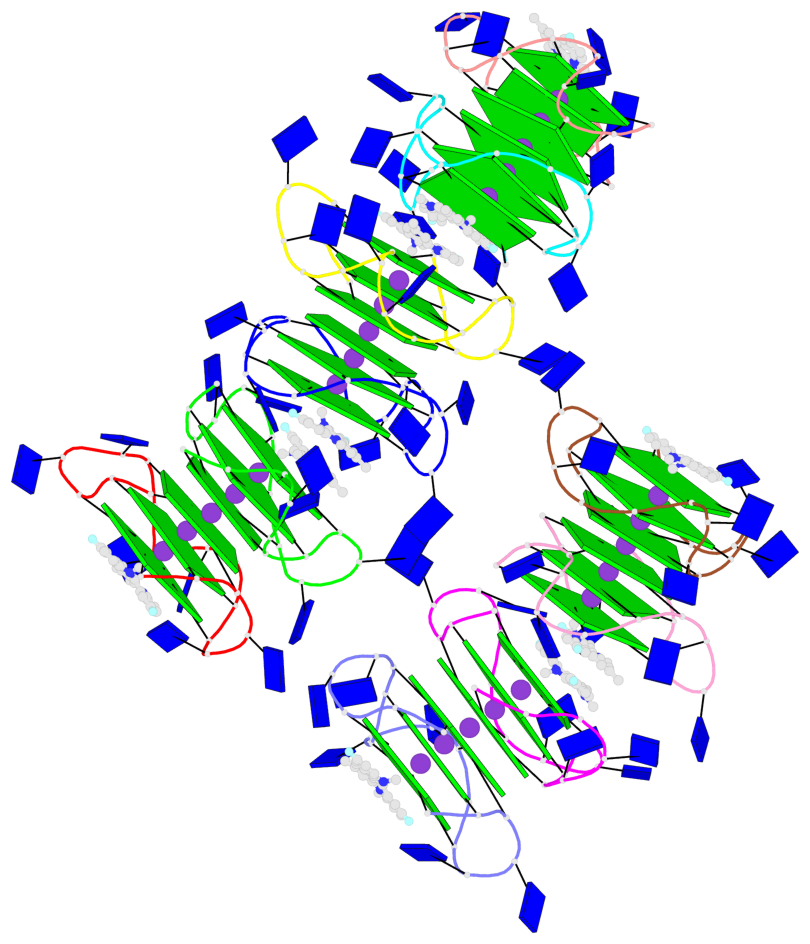
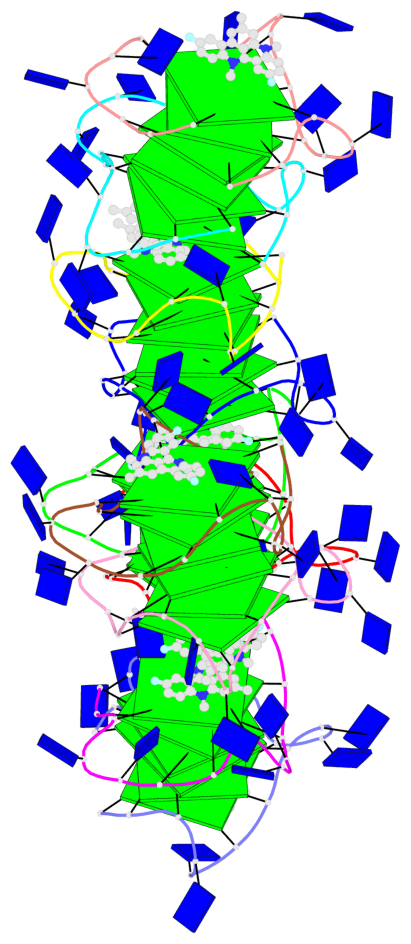
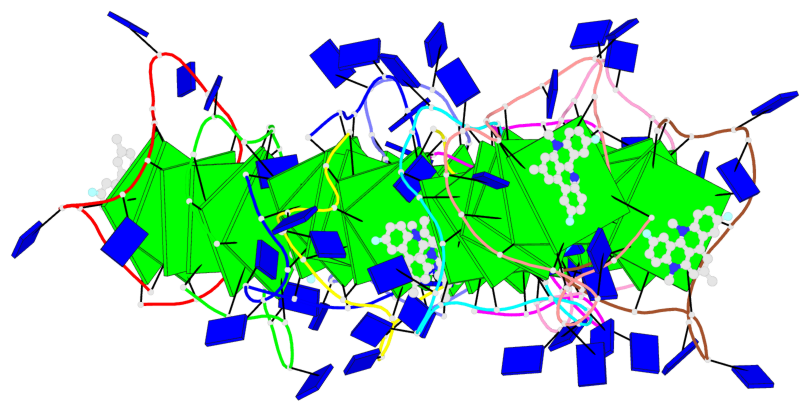
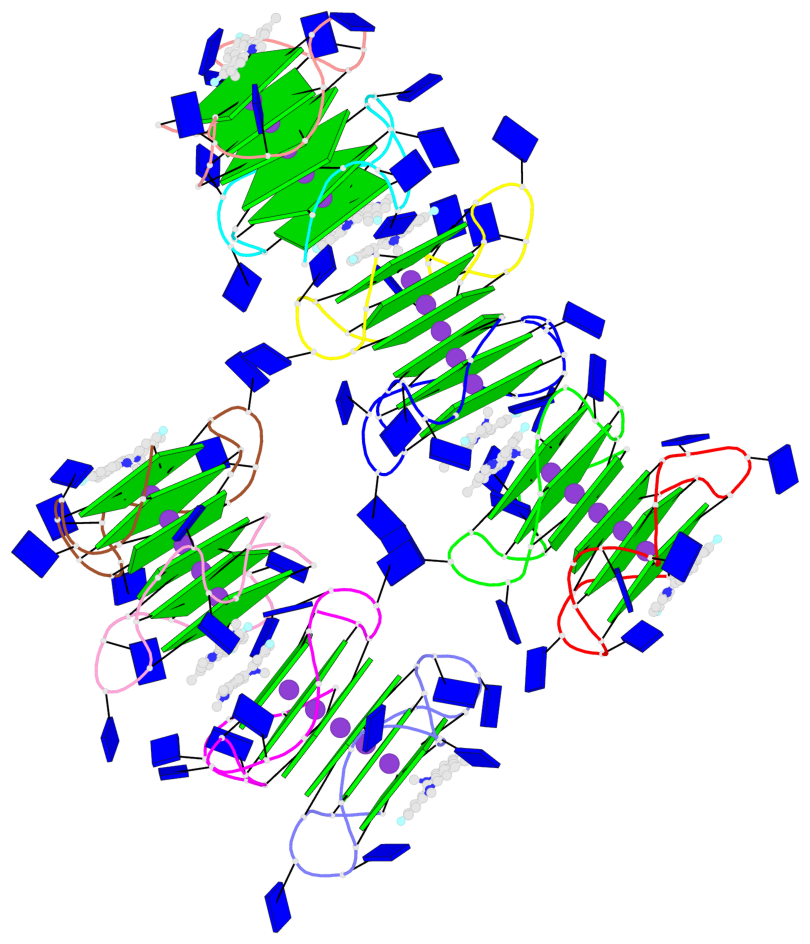
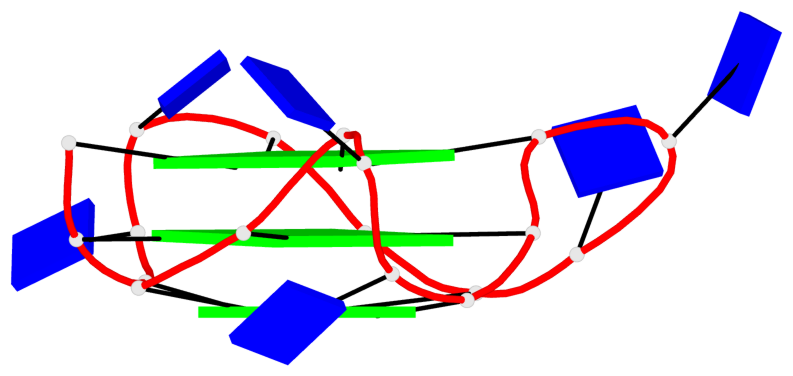
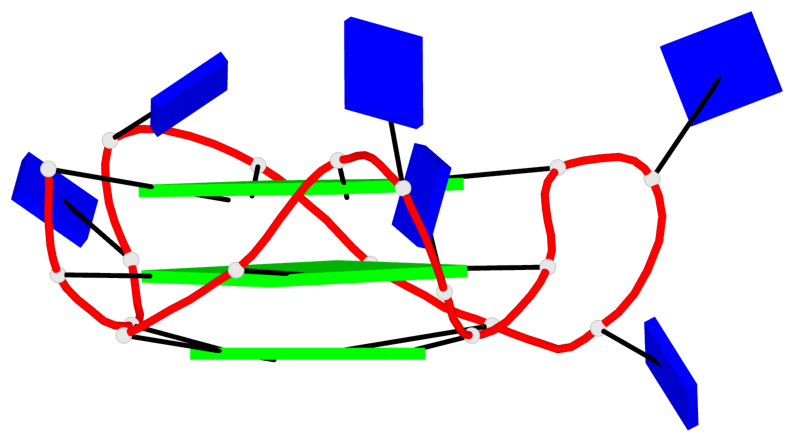
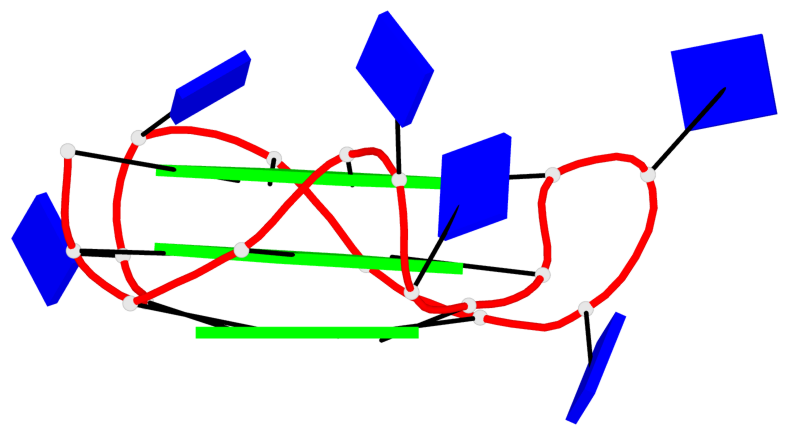
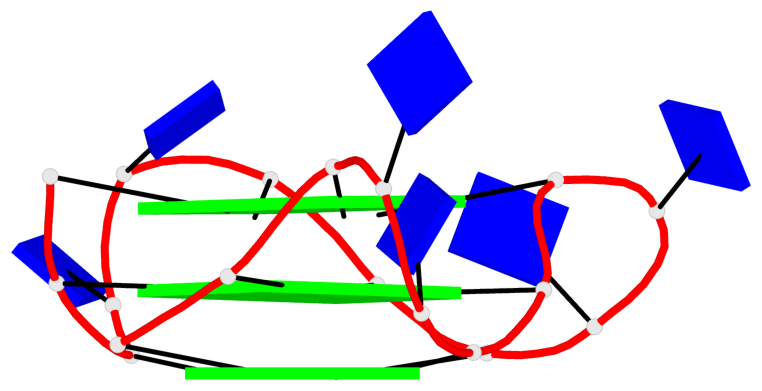
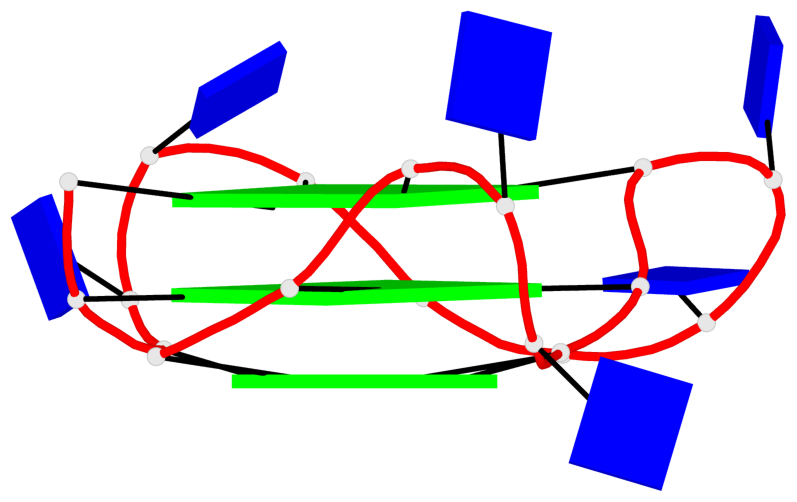
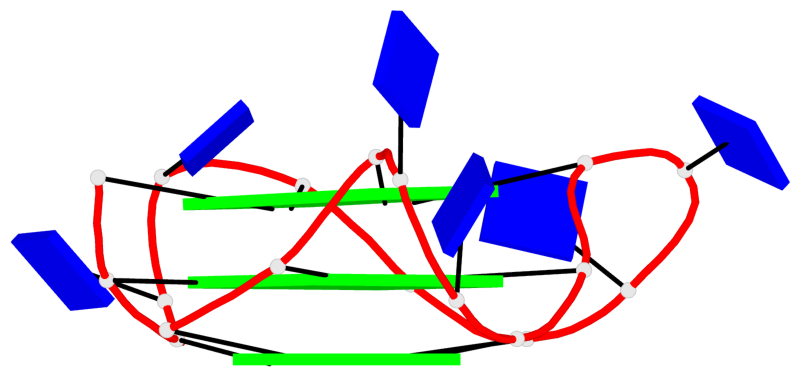
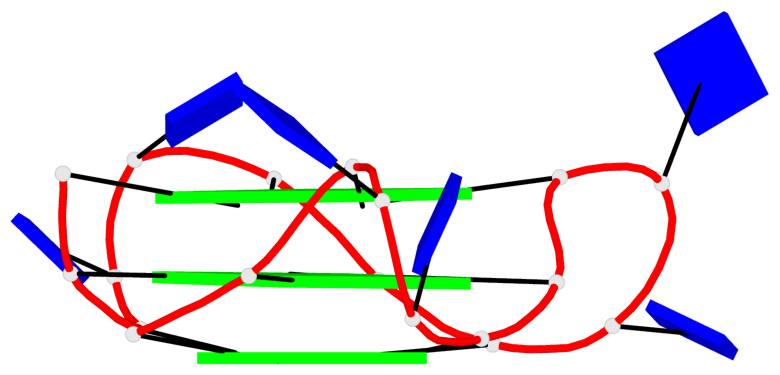
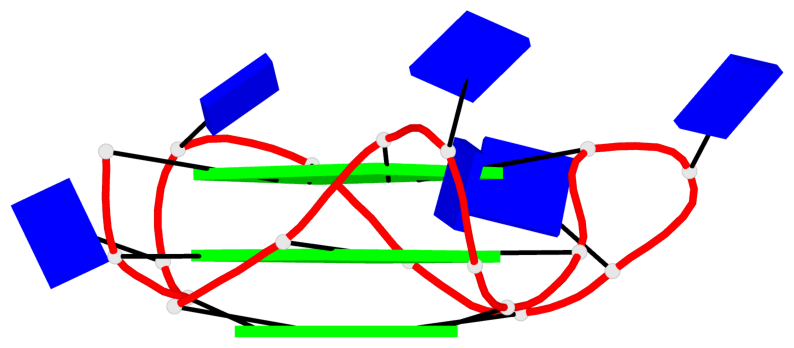
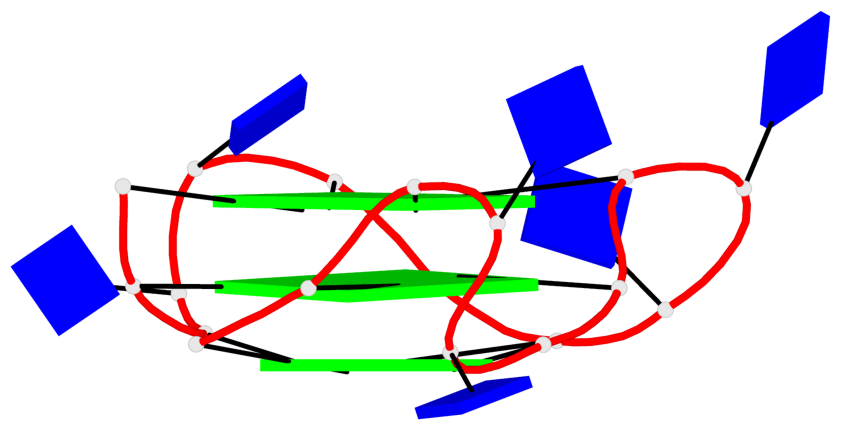
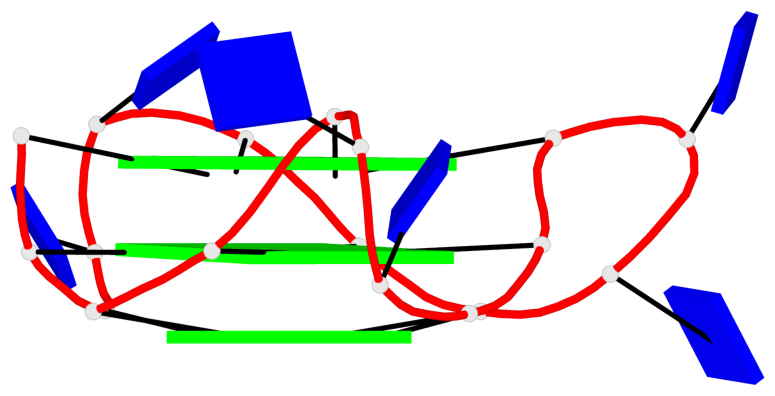
Detailed DSSR results for the G-quadruplex: PDB entry 8twh
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 8twh
- Class
- DNA
- Method
- X-ray (2.47 Å)
- Summary
- Crystal structure of (gggtt)3ggg g-quadruplex in complex with small molecule ligand rhps4
- Reference
- Lam G, Martin KN, Yatsunyk LA: "Crystal structure of (GGGTT)3GGG G-quadruplex in complex with small molecule ligand RHPS4."
- Abstract
- G4 notes
- 30 G-tetrads, 5 G4 helices, 10 G4 stems, 5 G4 coaxial stacks, 3(-P-P-P), parallel(4+0), UUUU, coaxial interfaces: 5'/5'
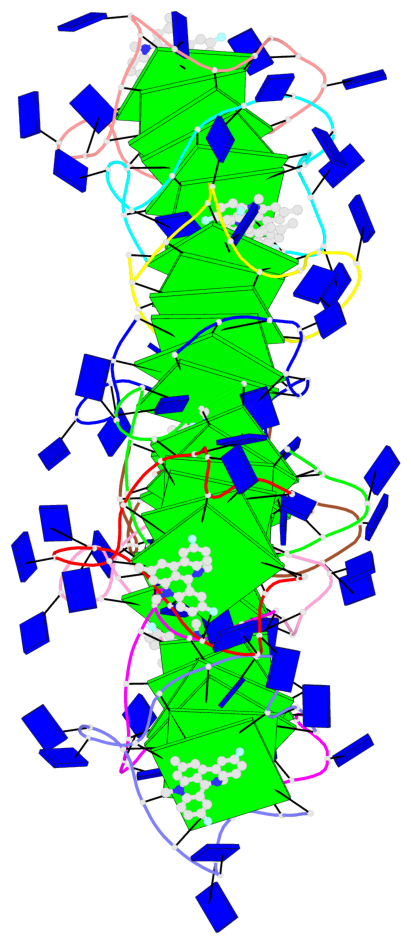
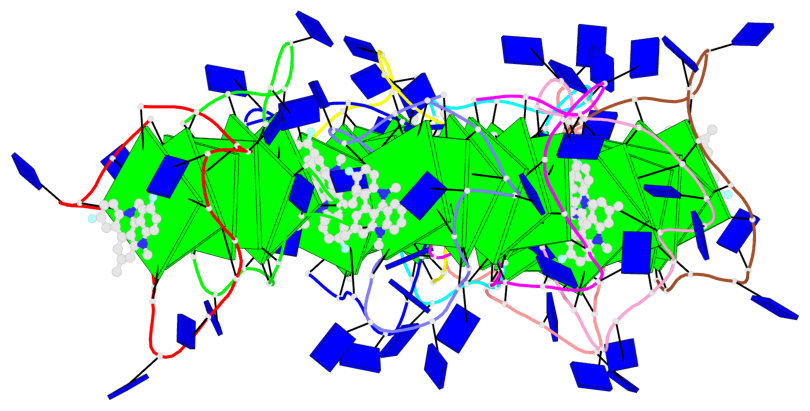
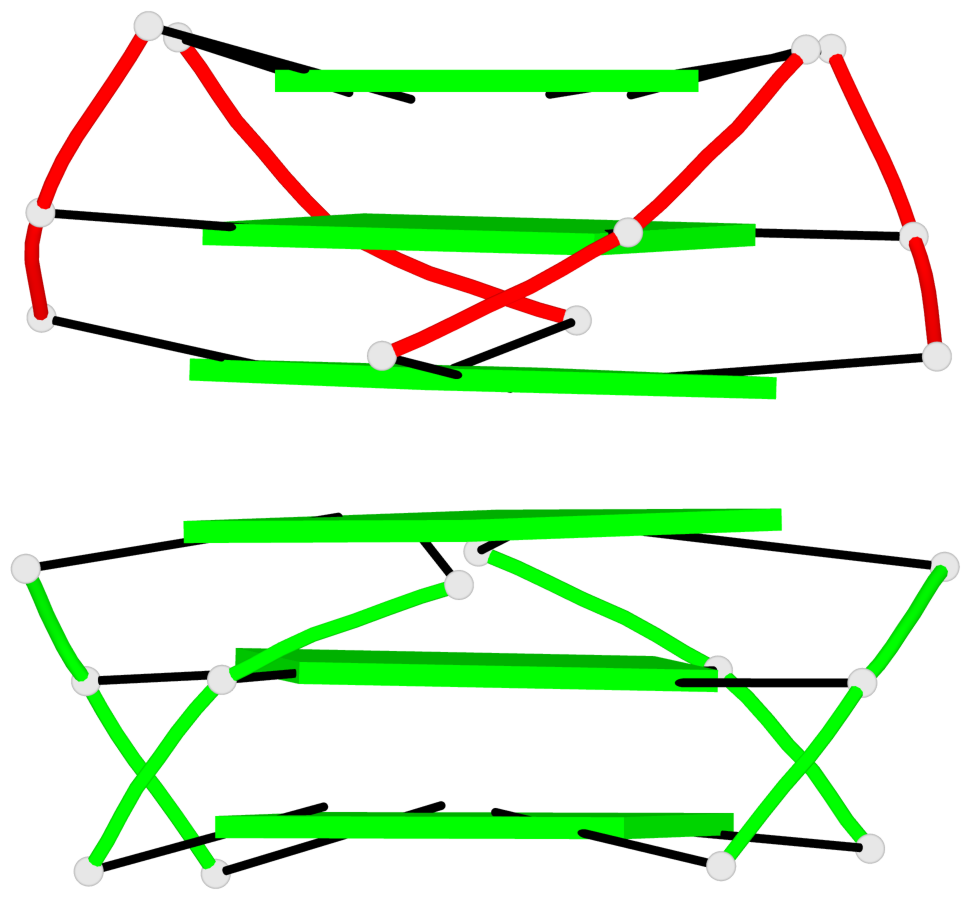
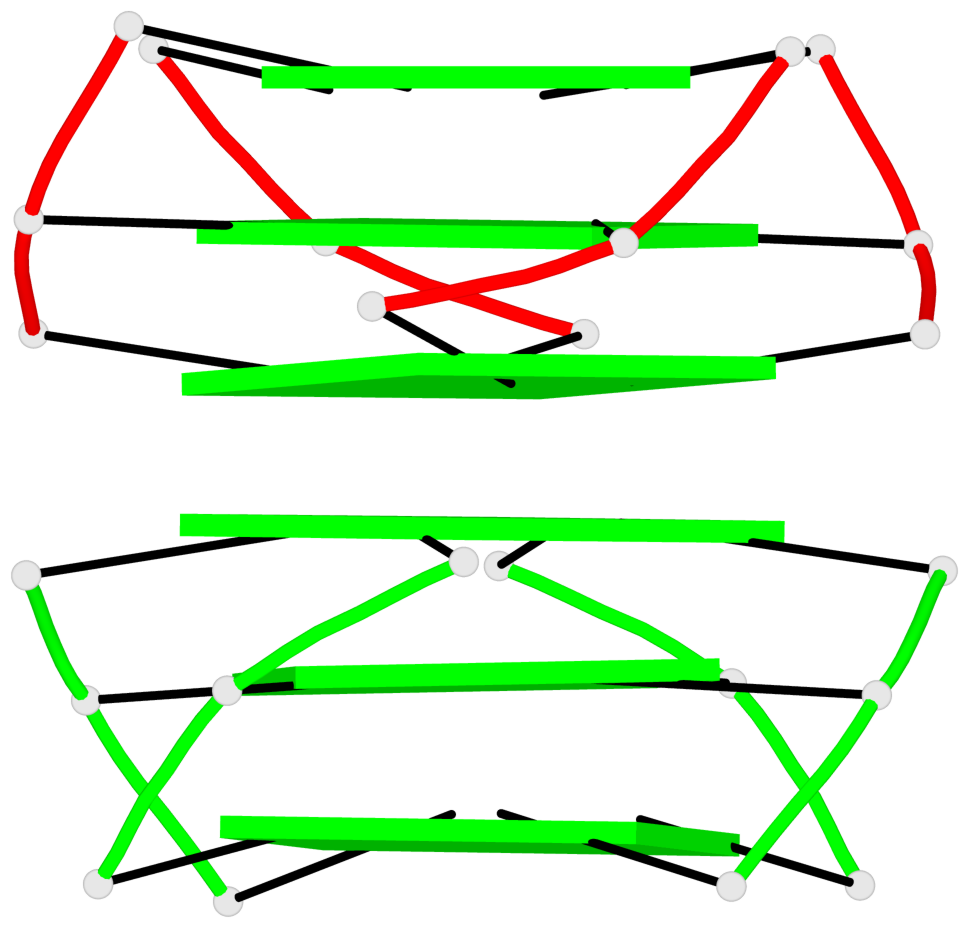
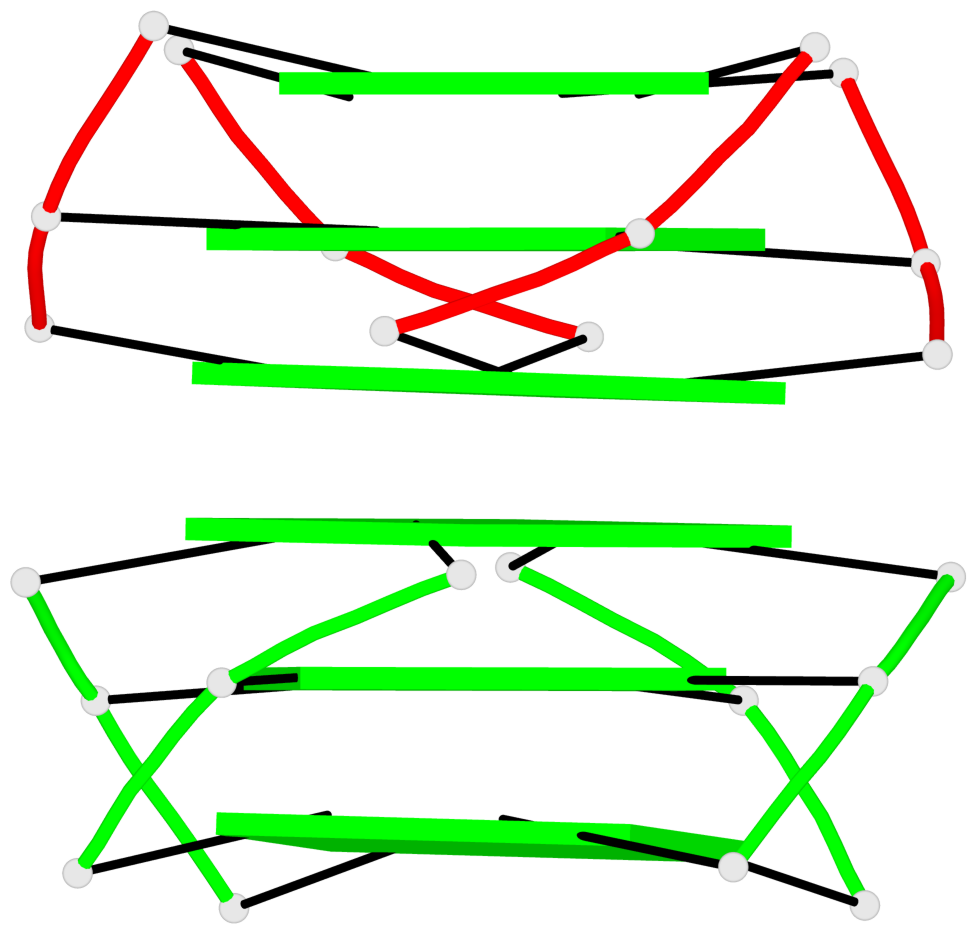
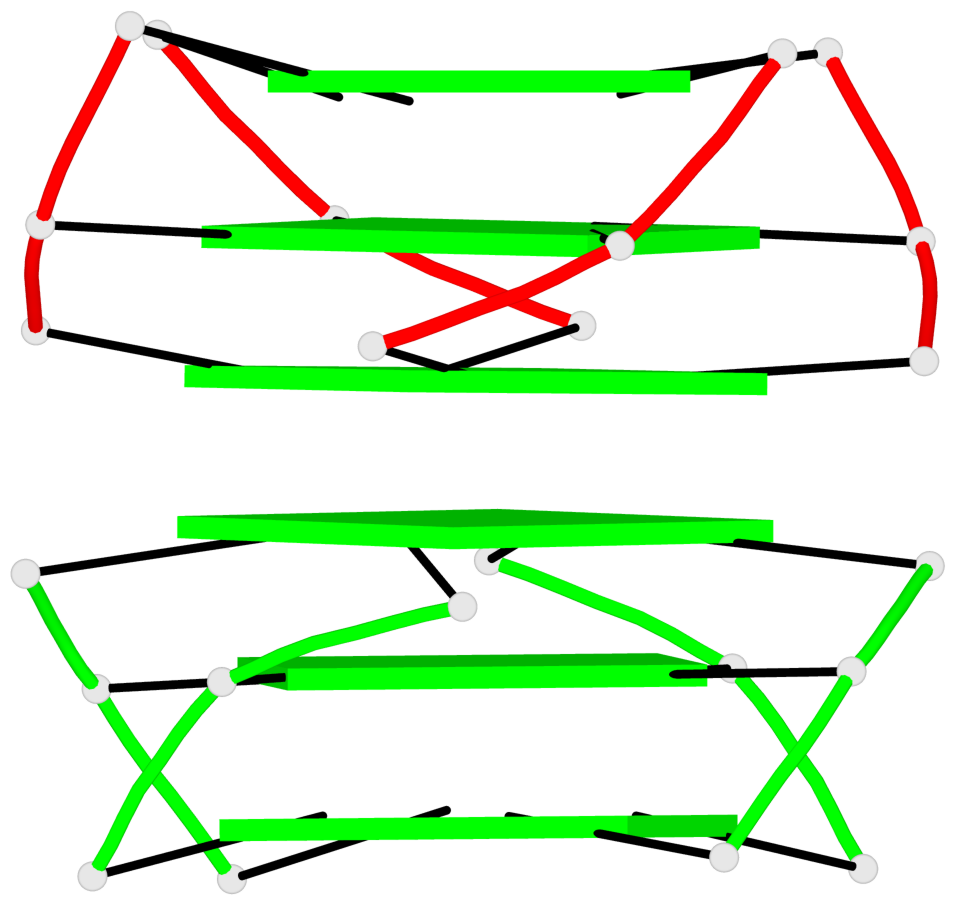
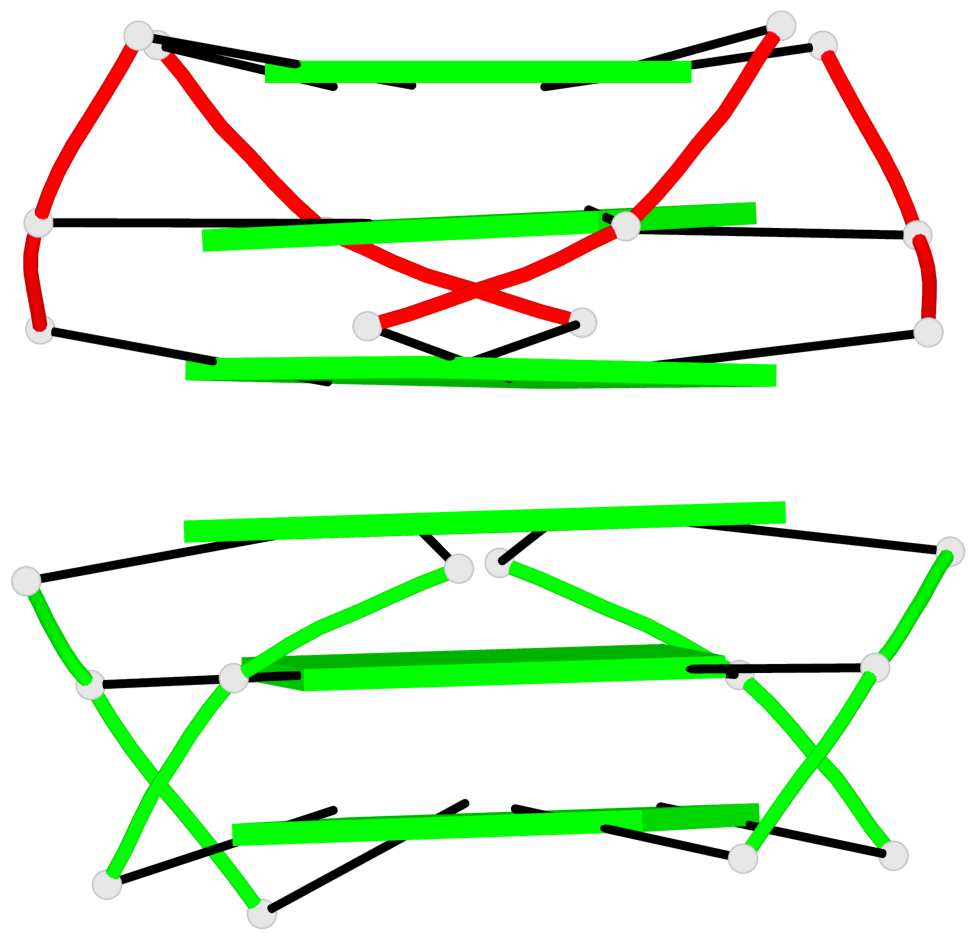
Base-block schematics in six views
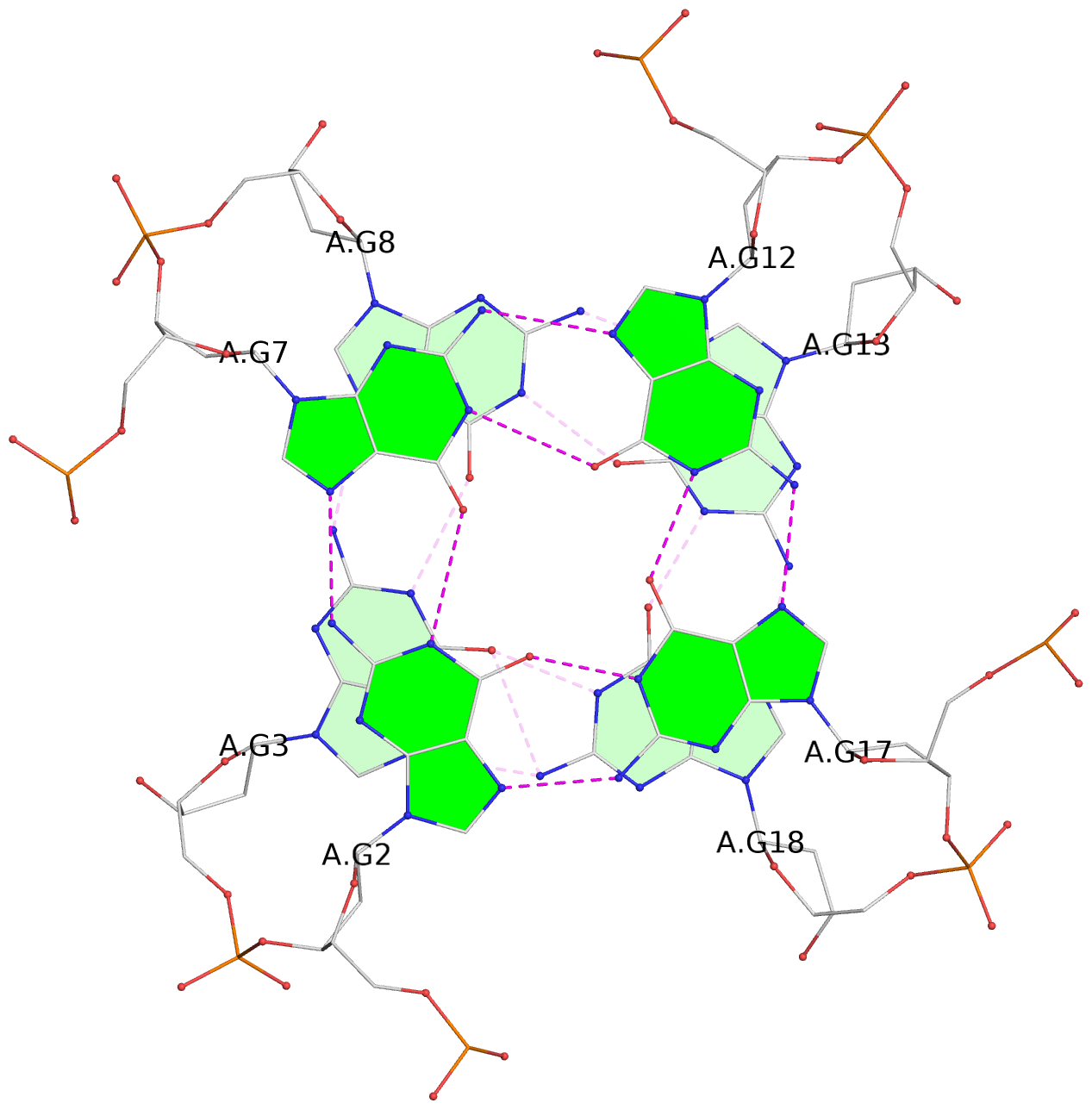
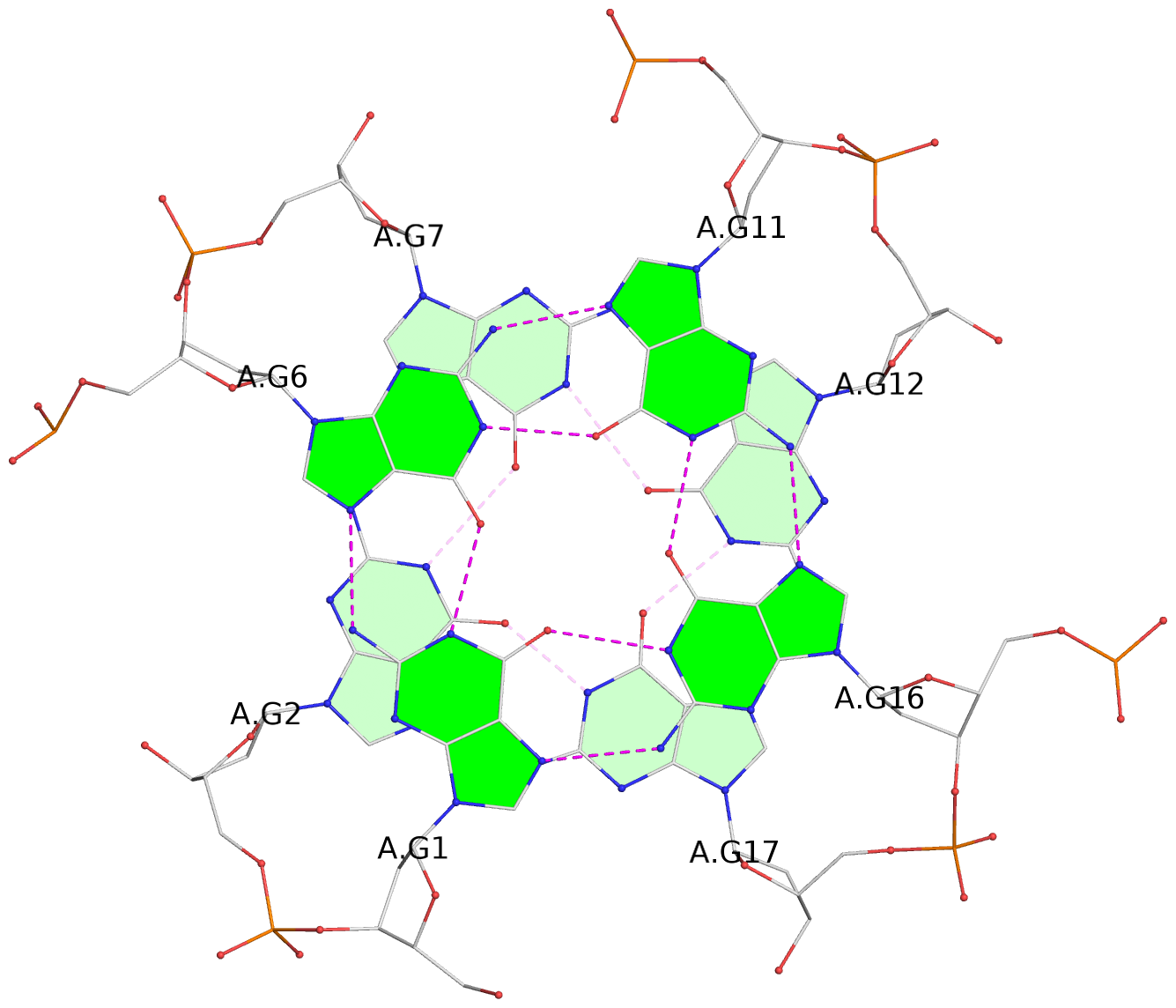
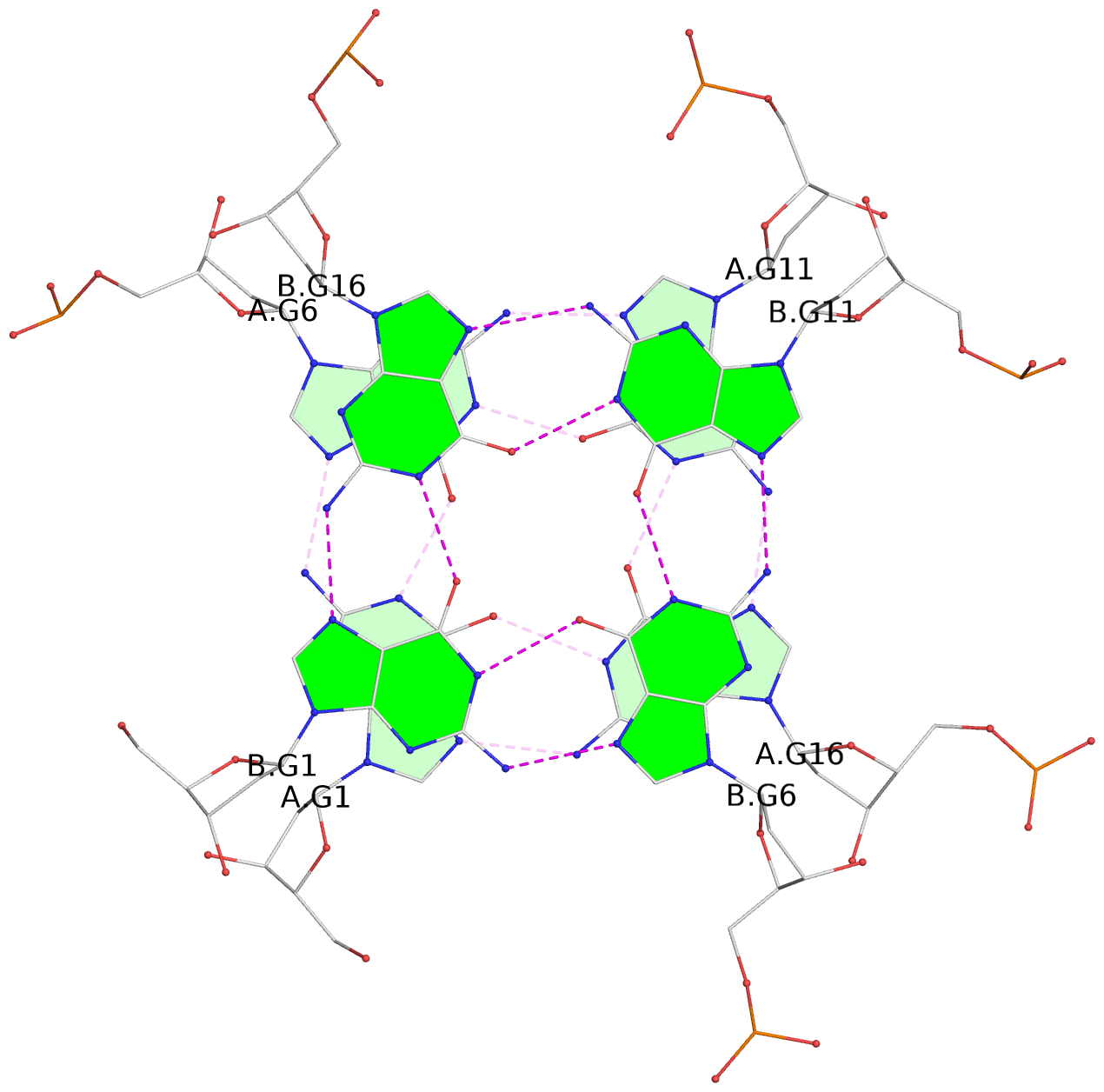
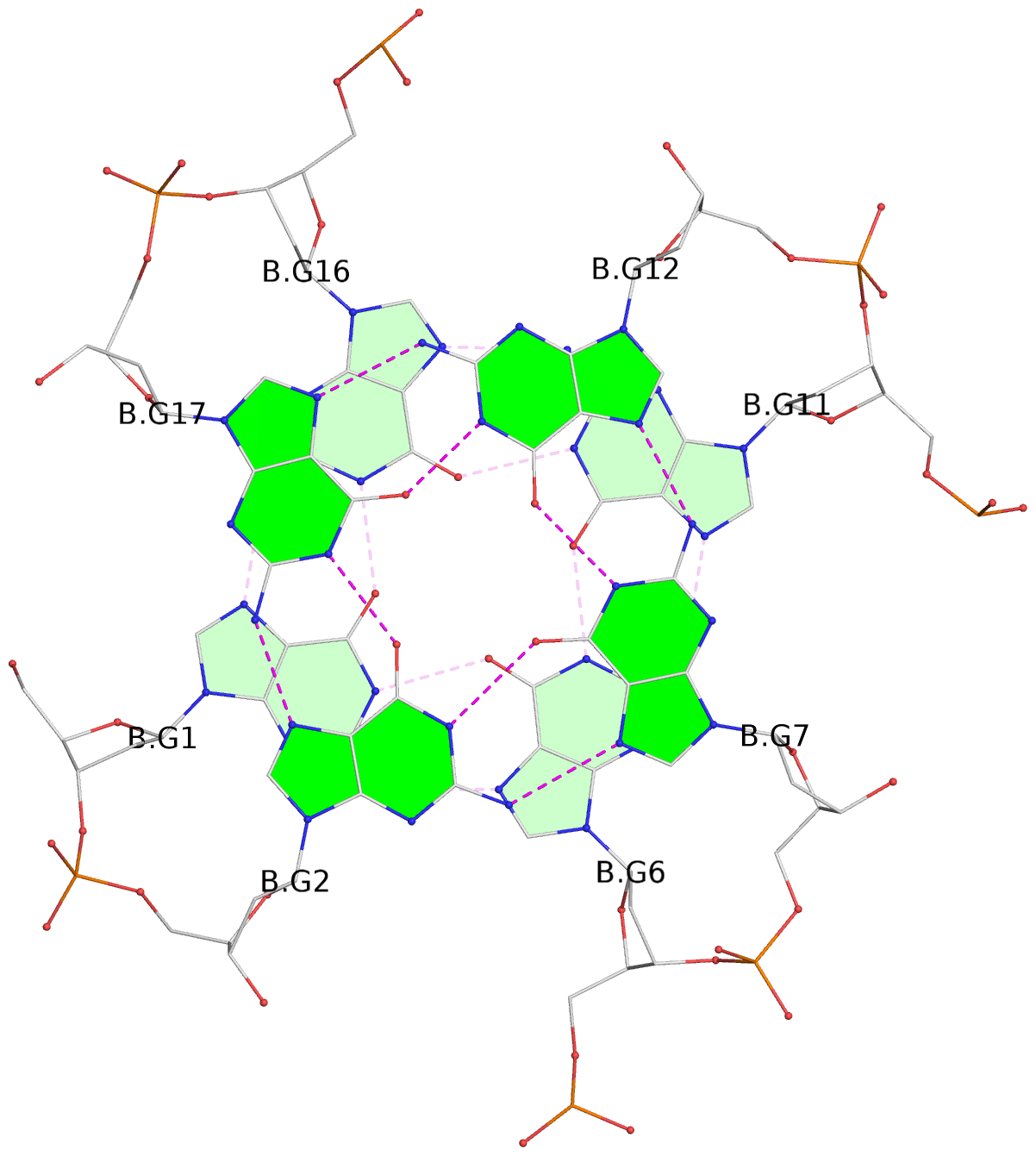
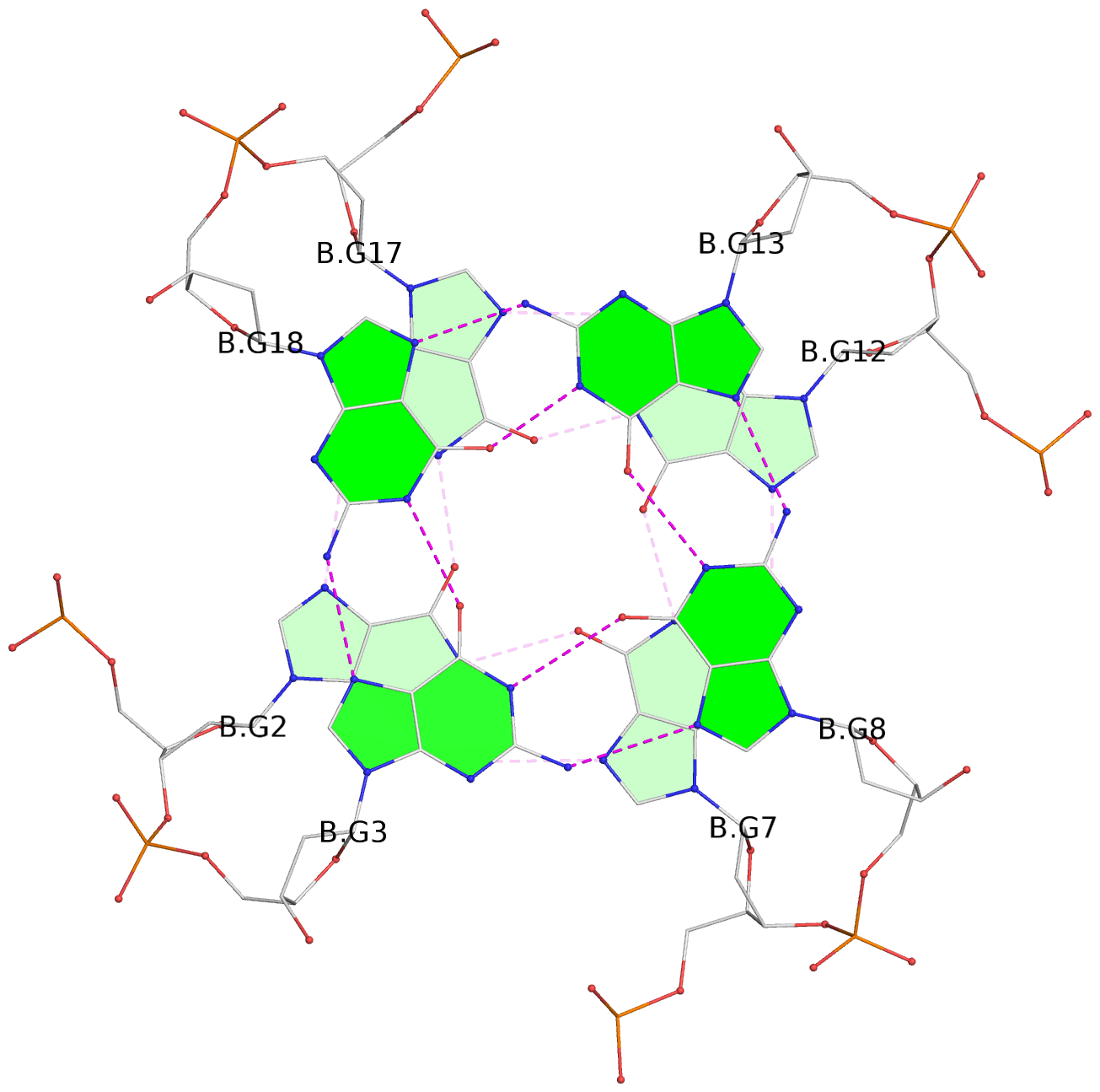
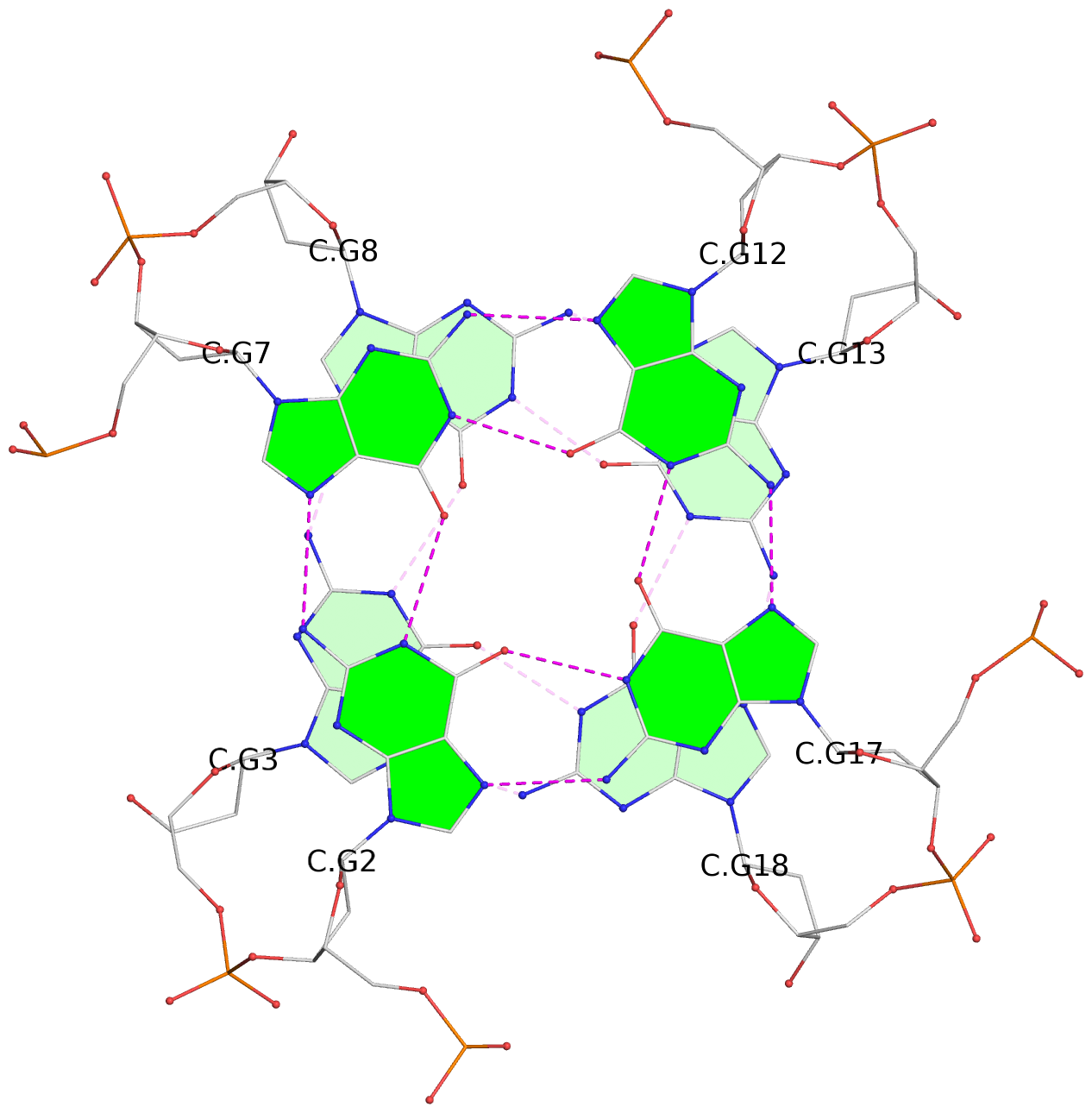
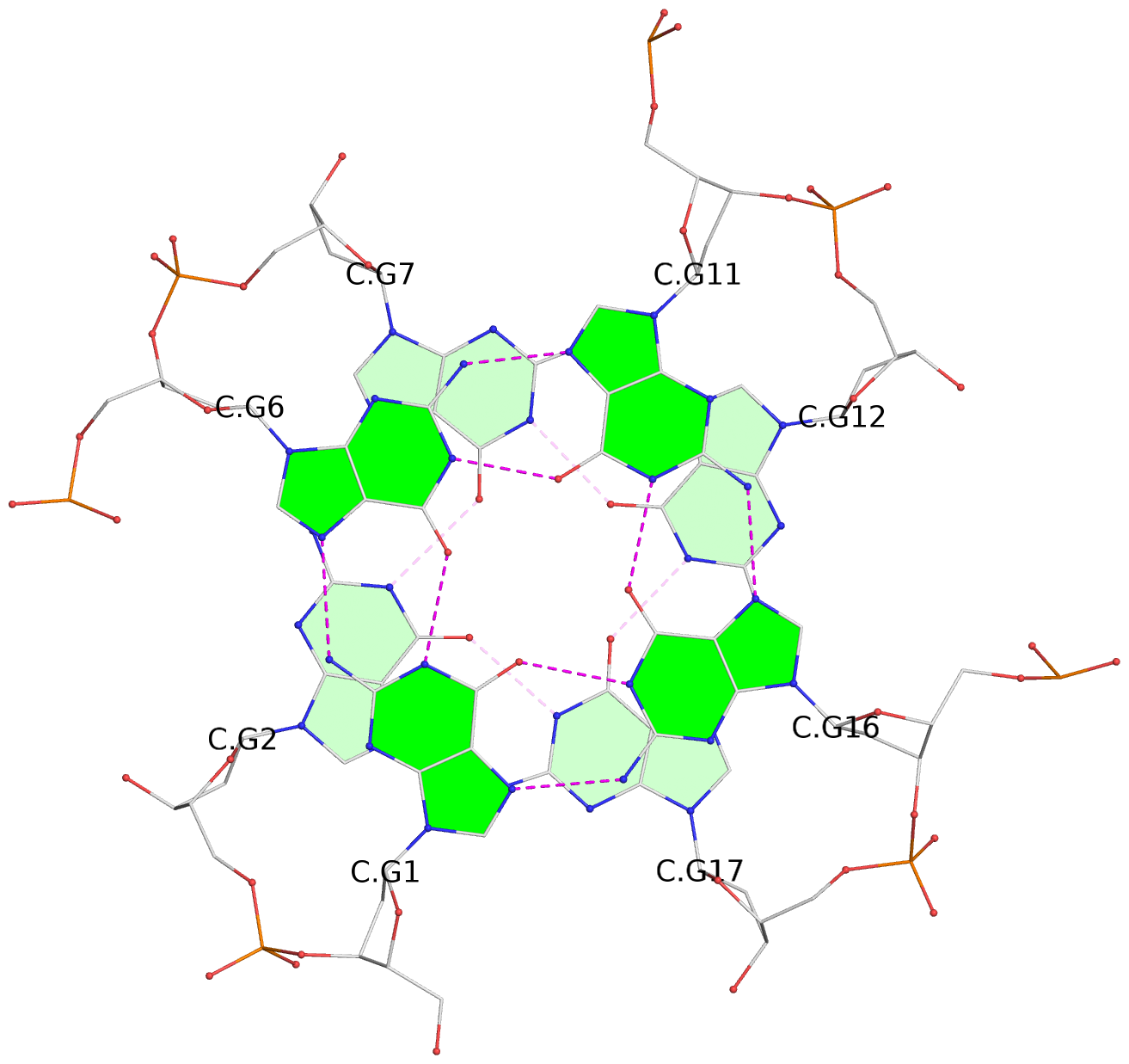
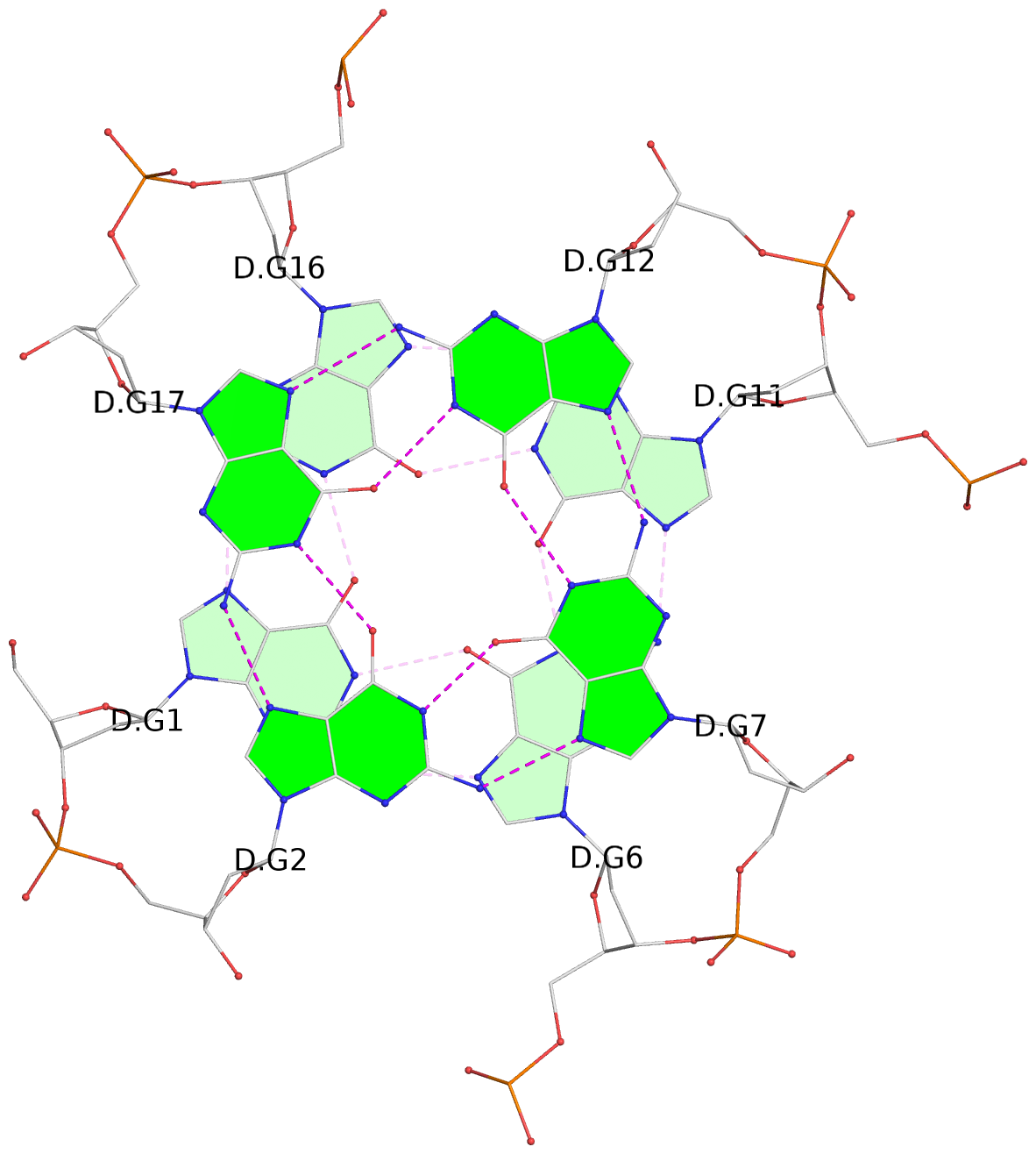
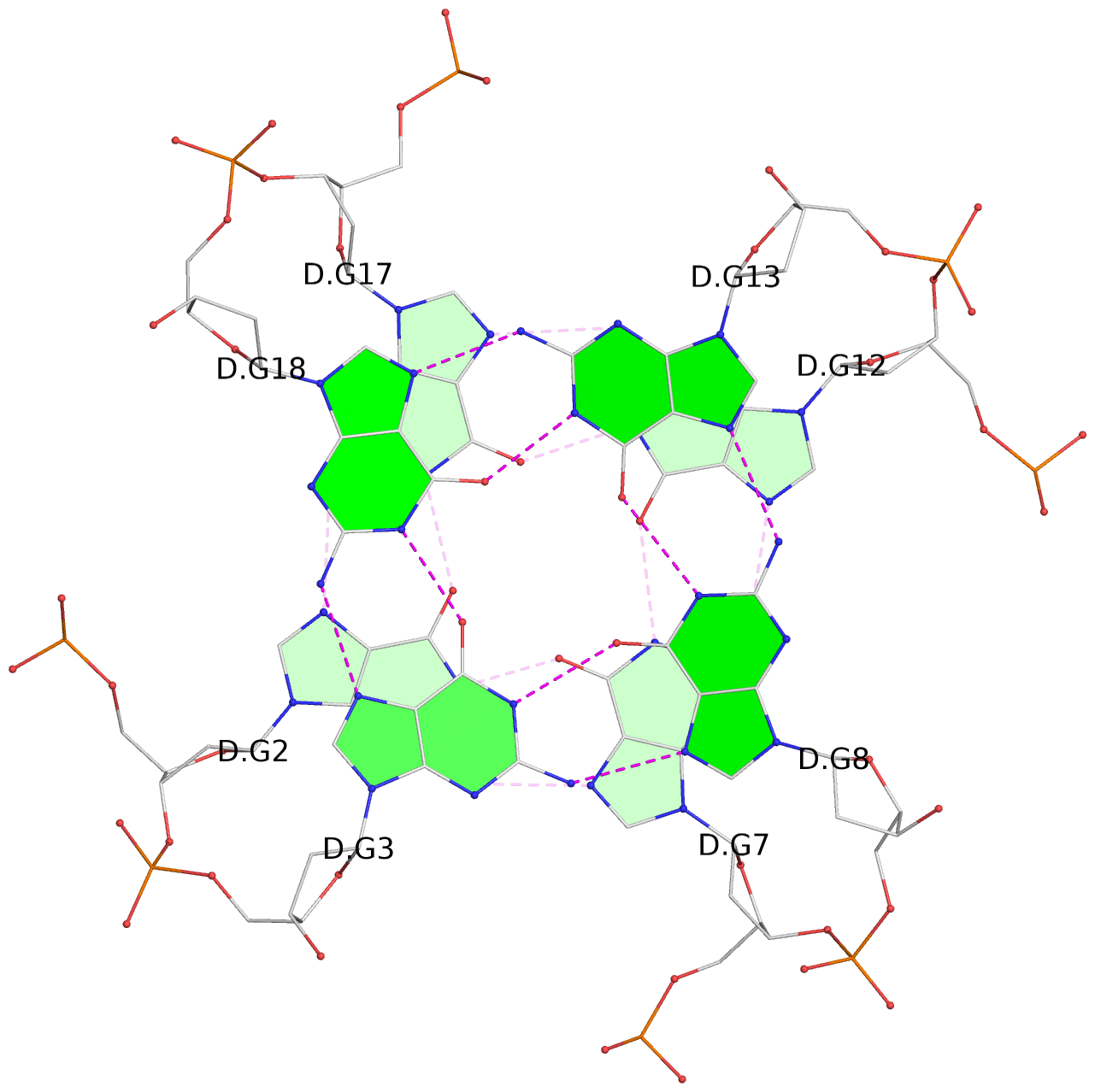
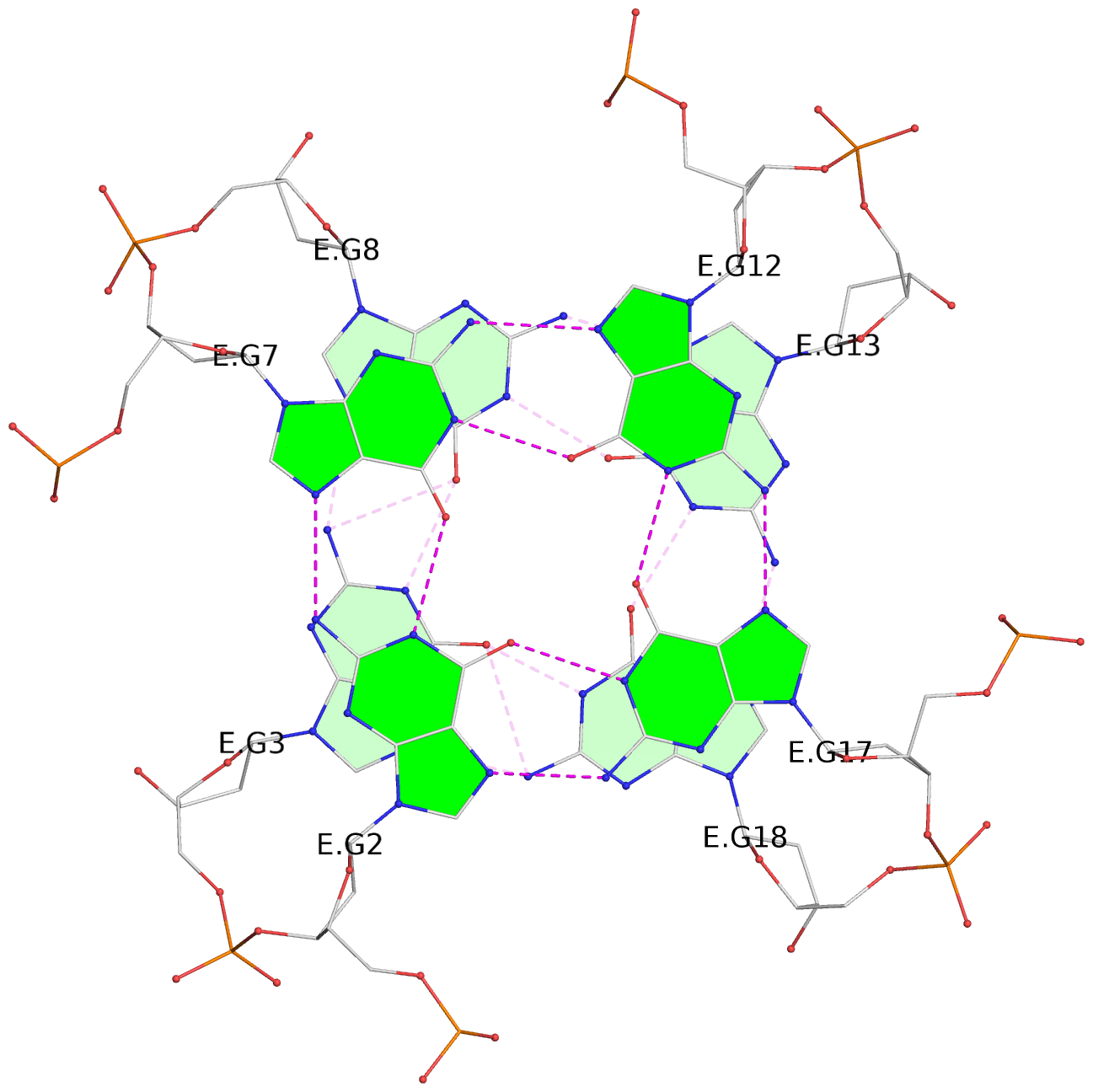
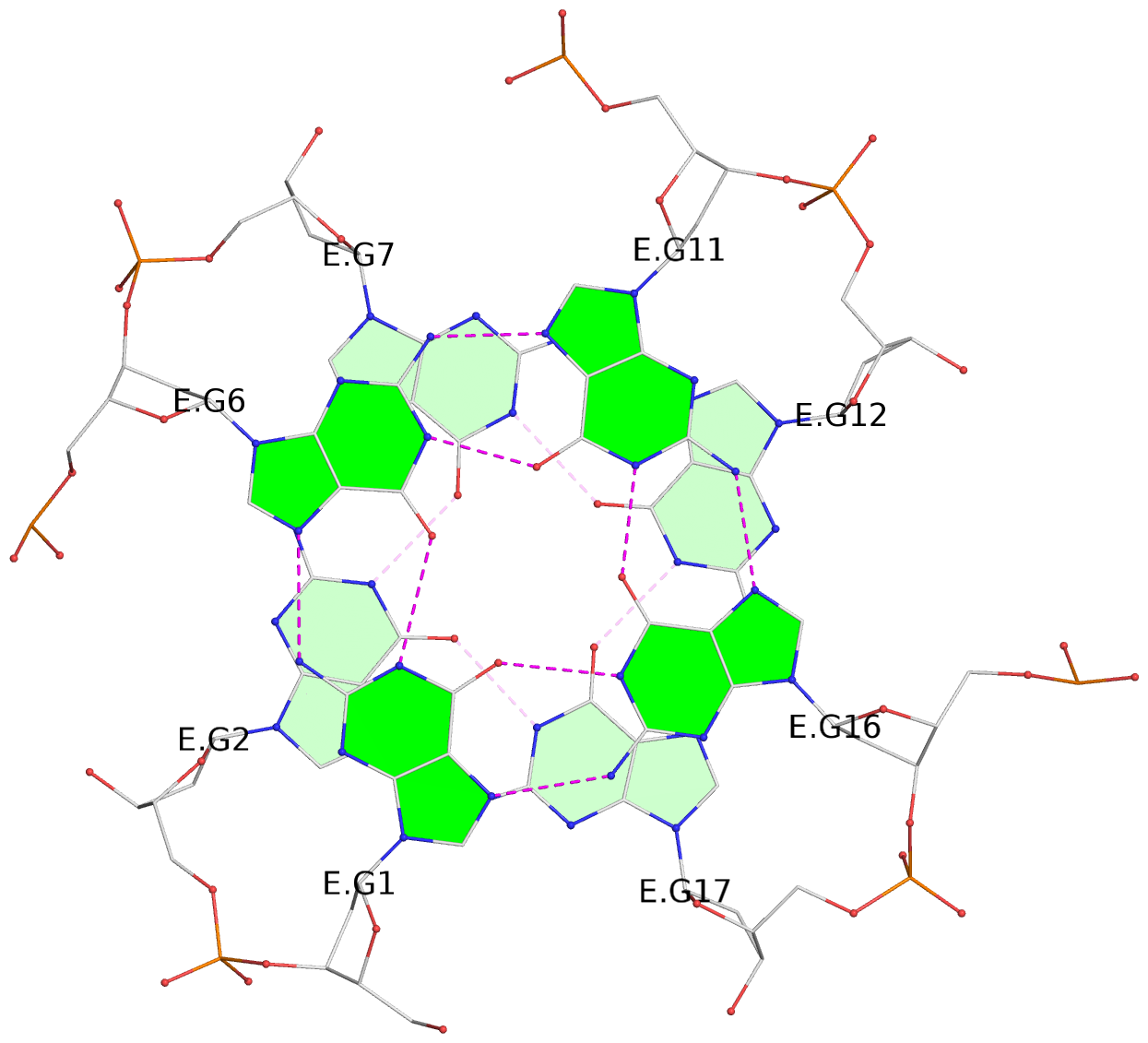
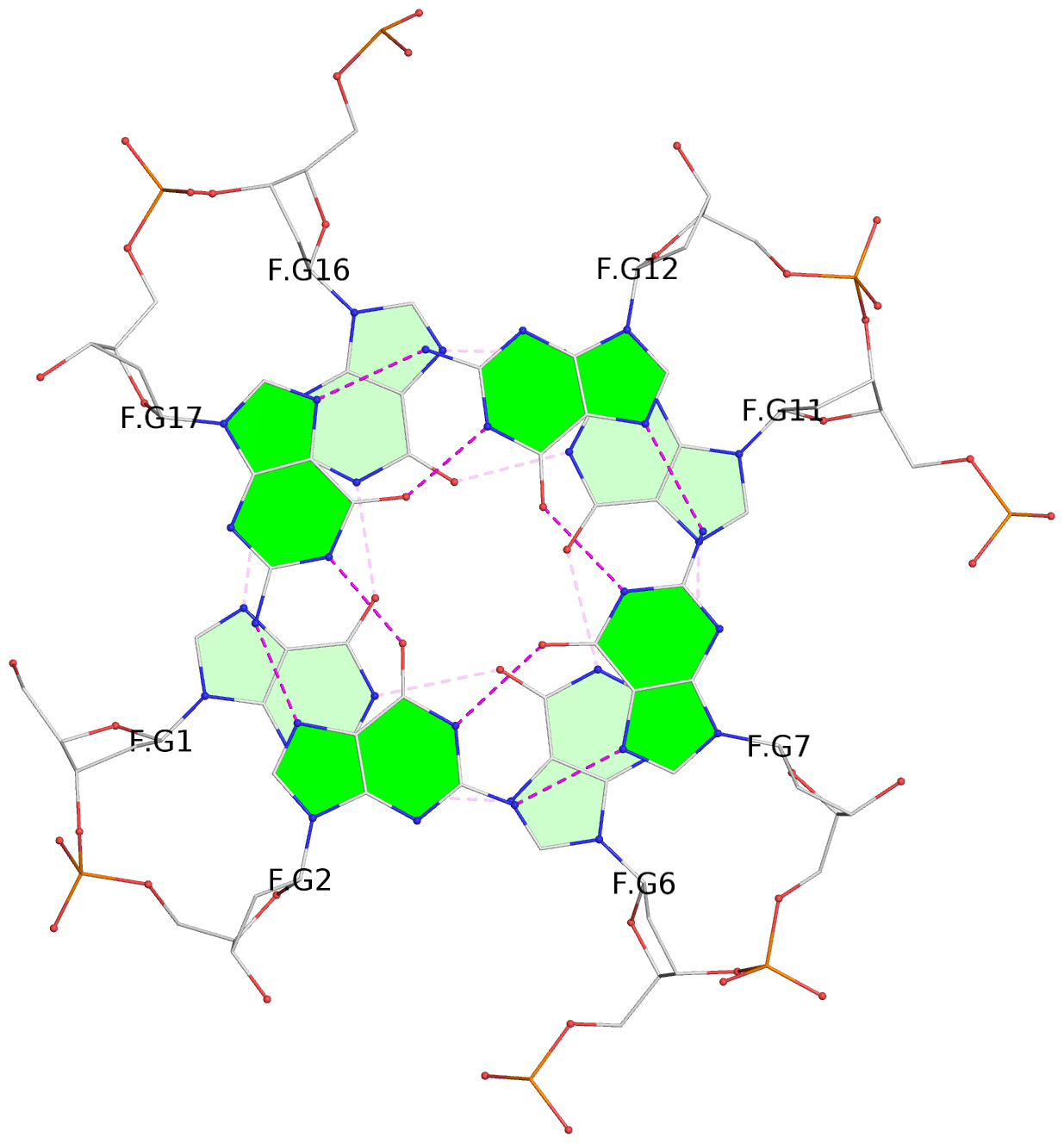
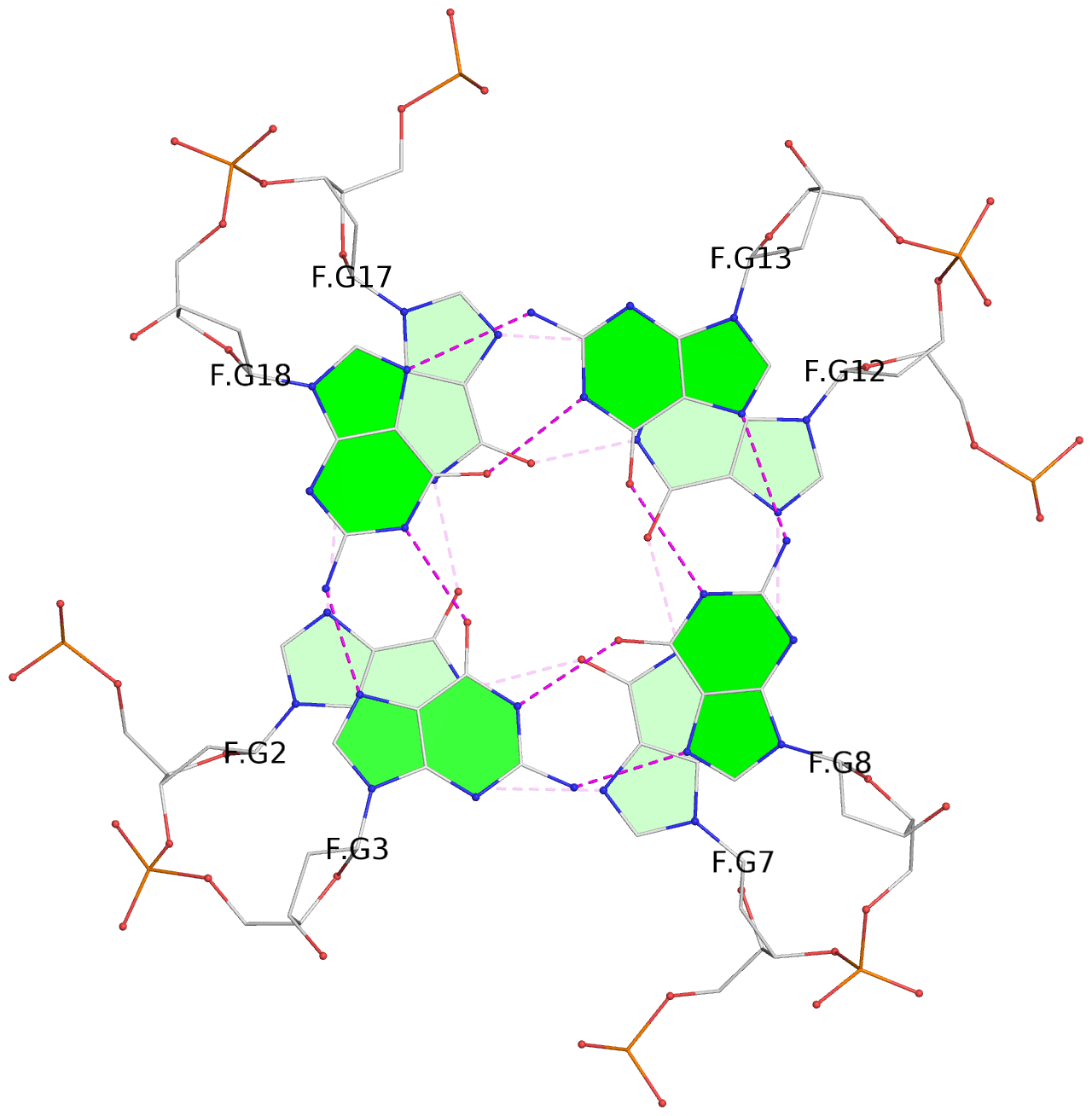
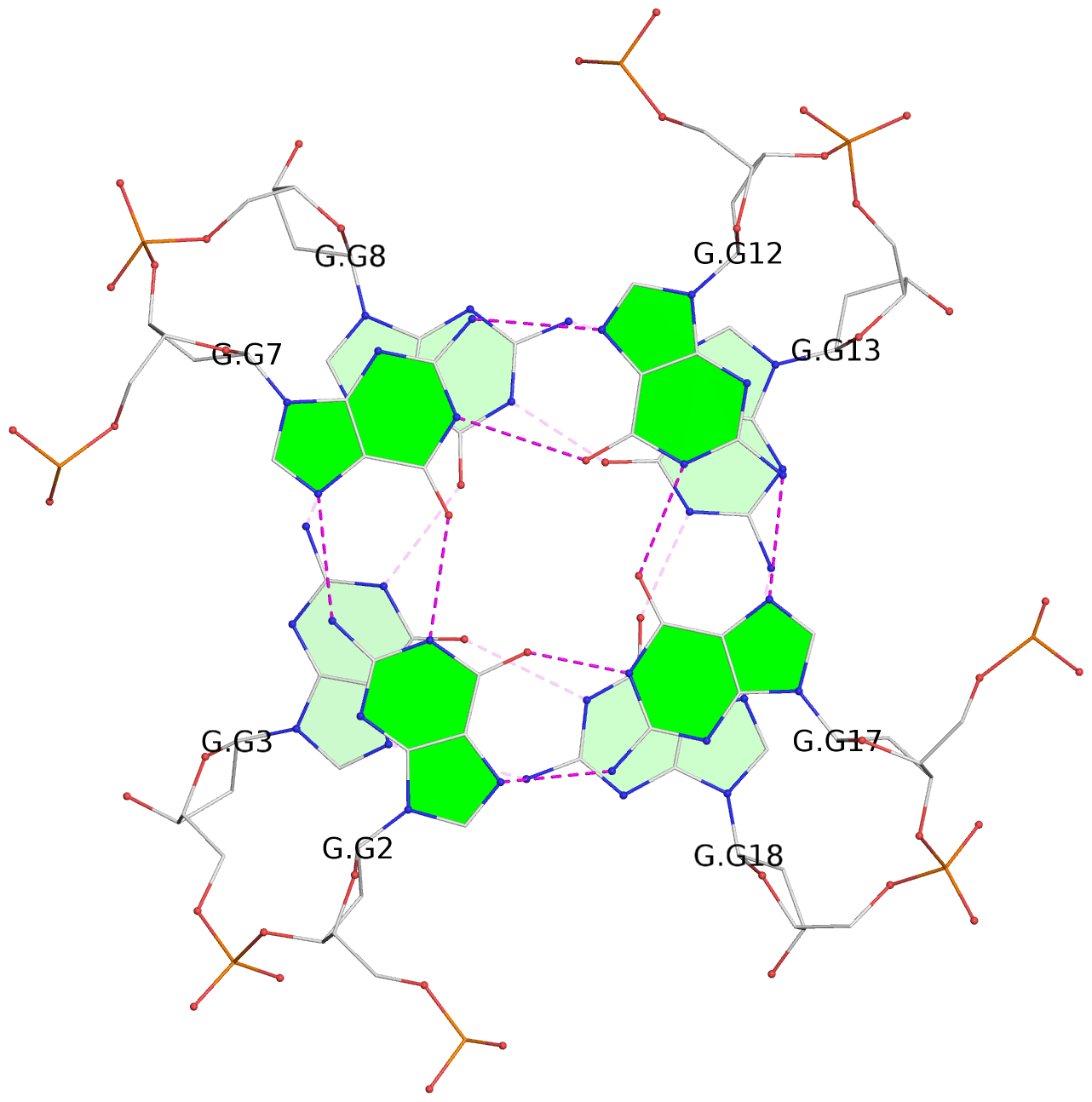
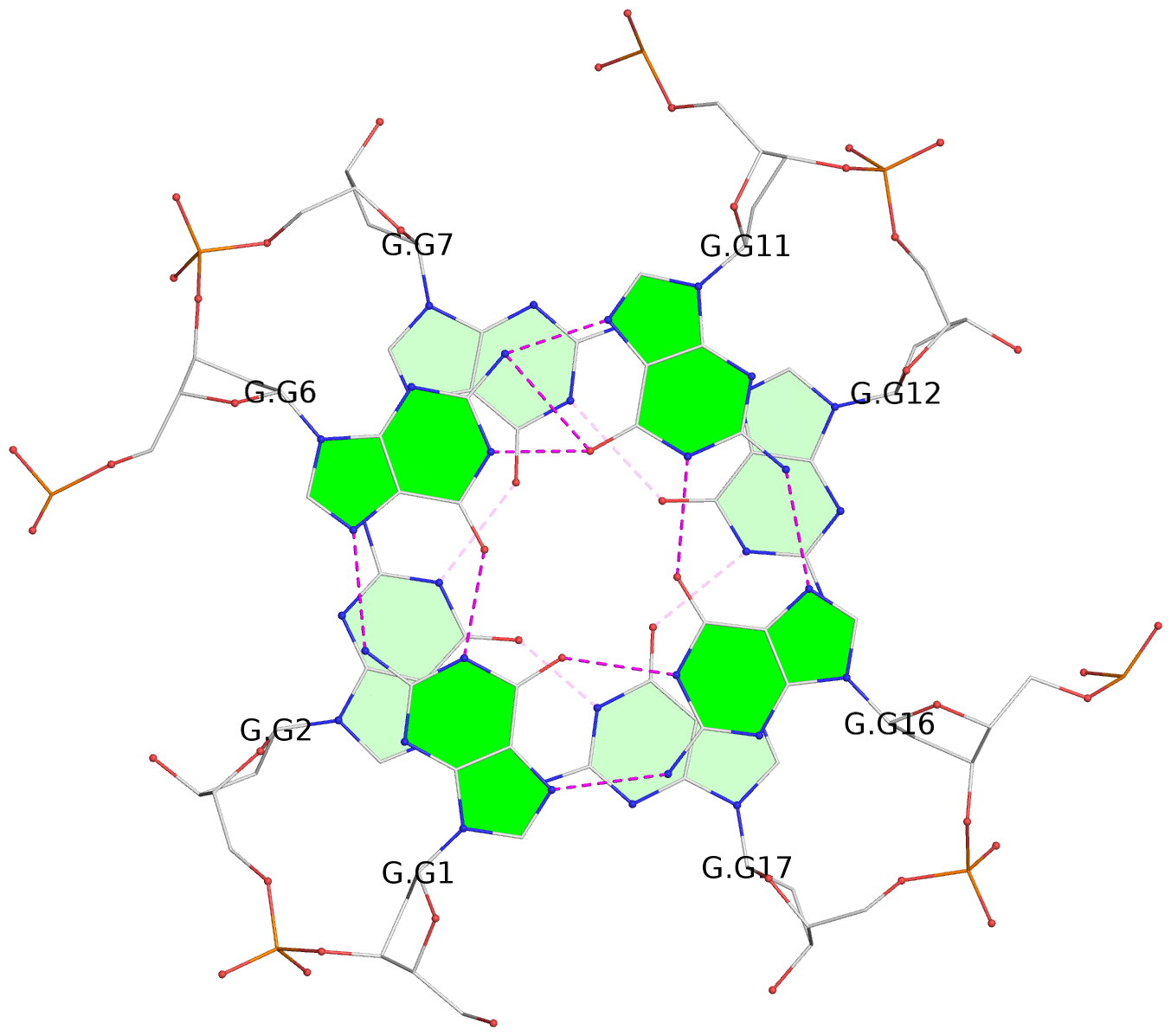
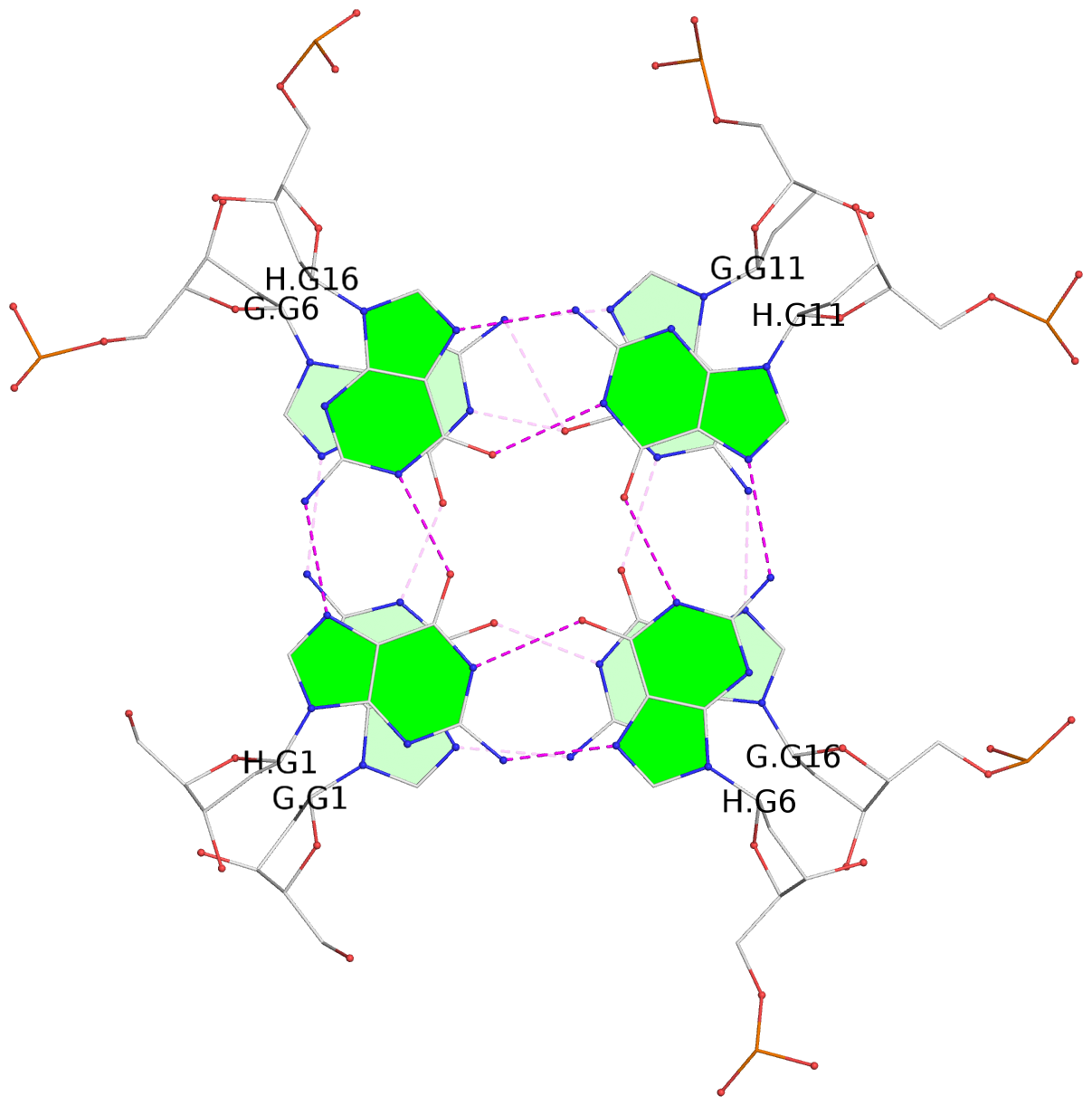
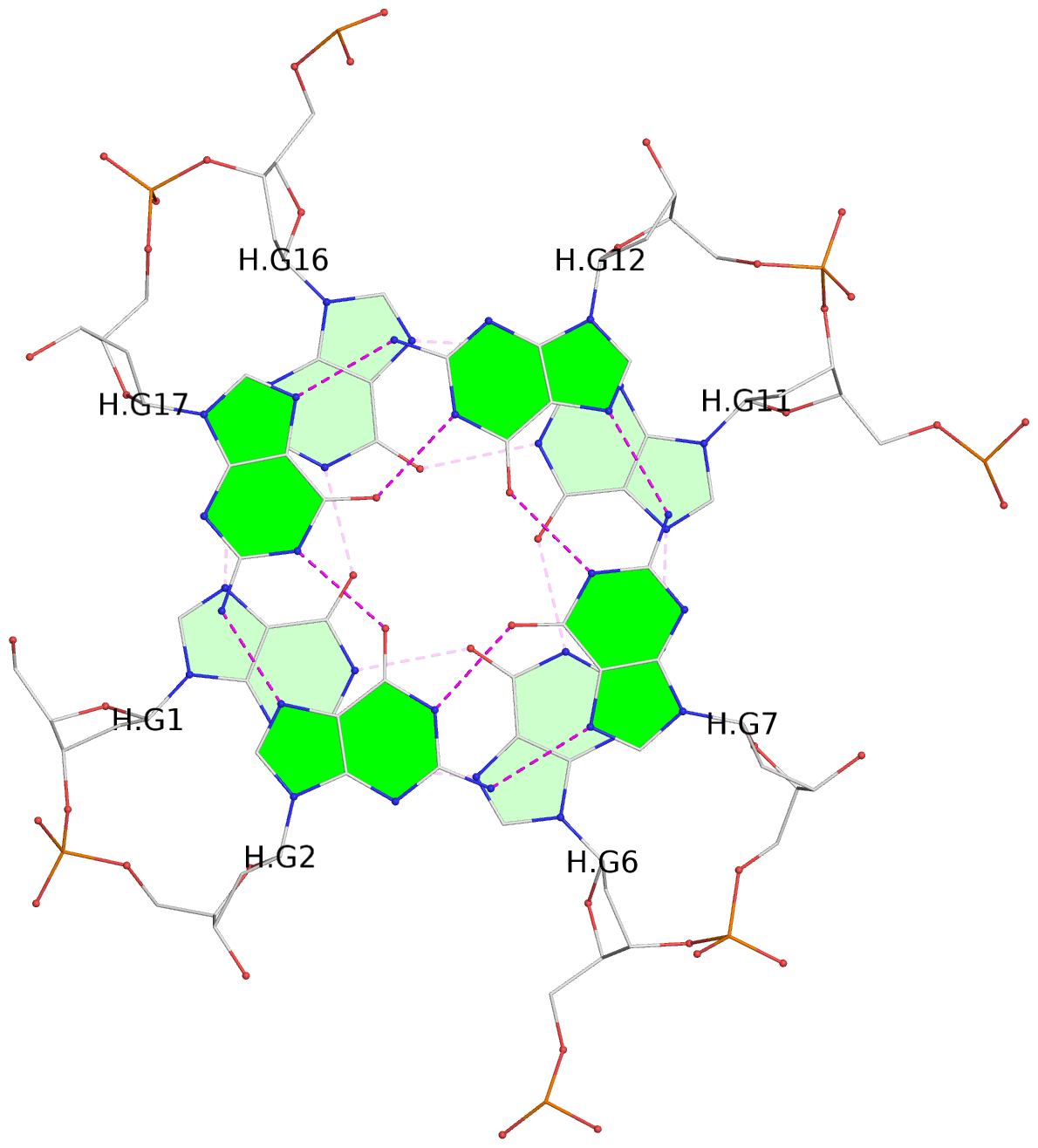
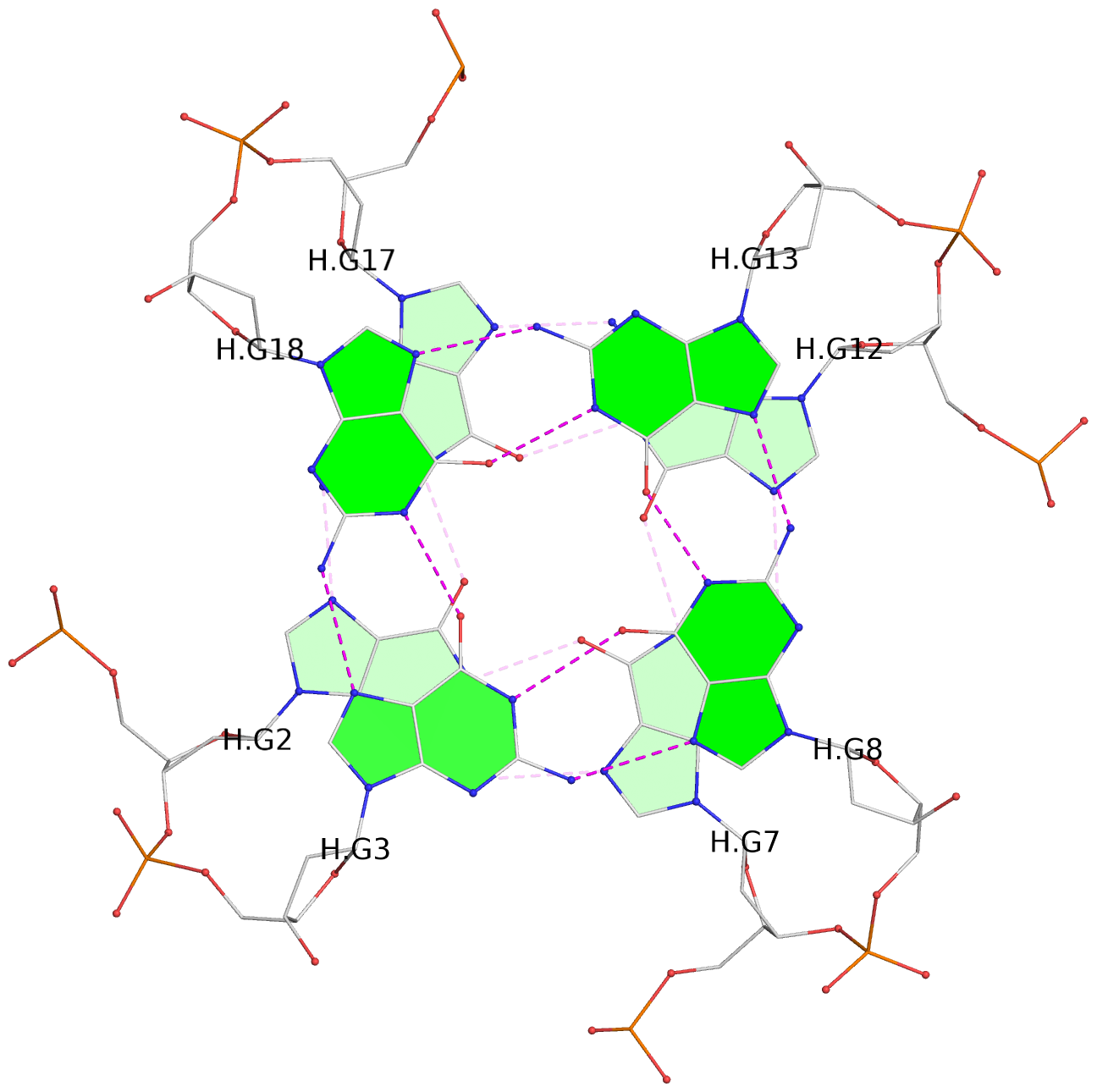
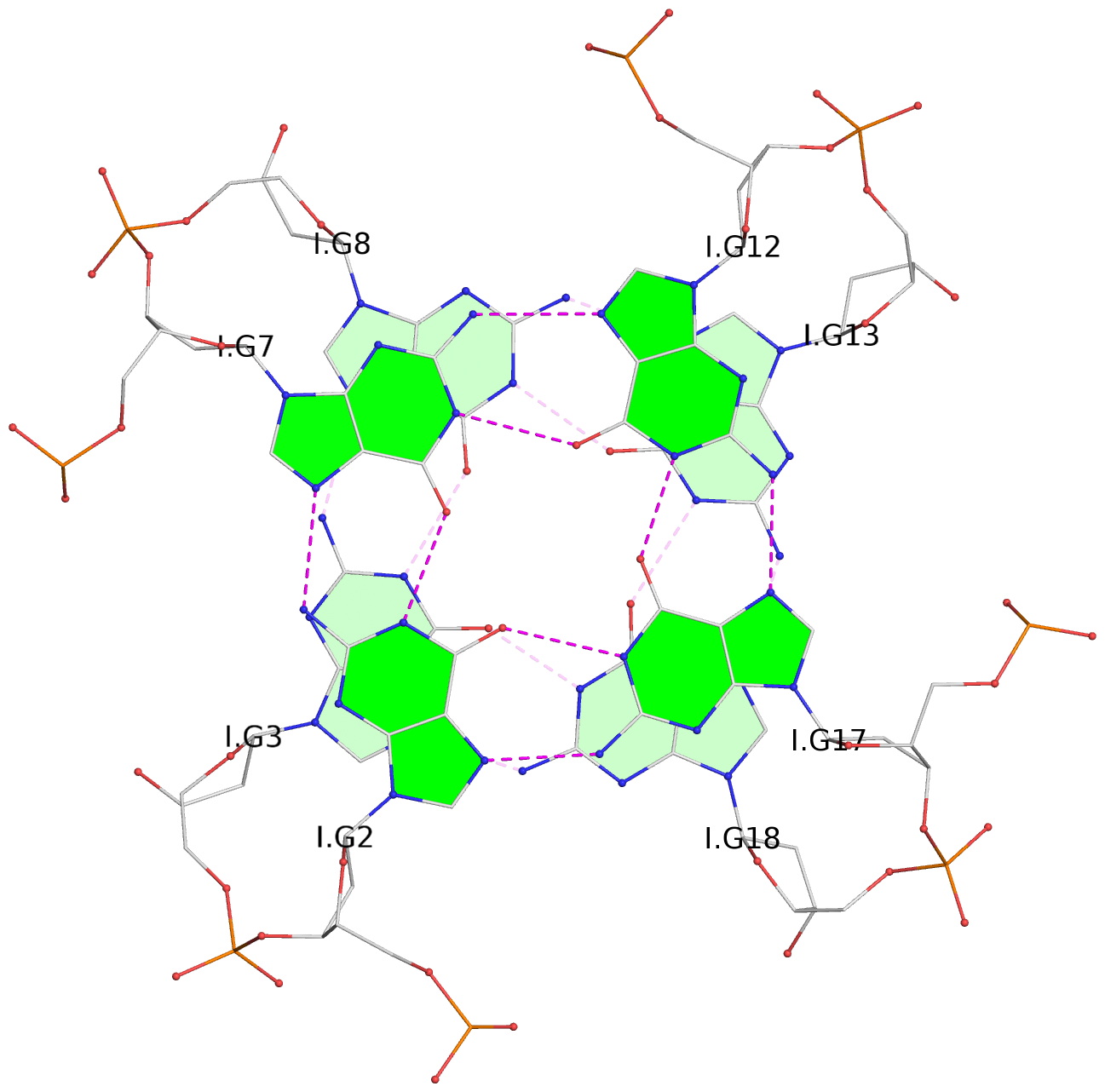
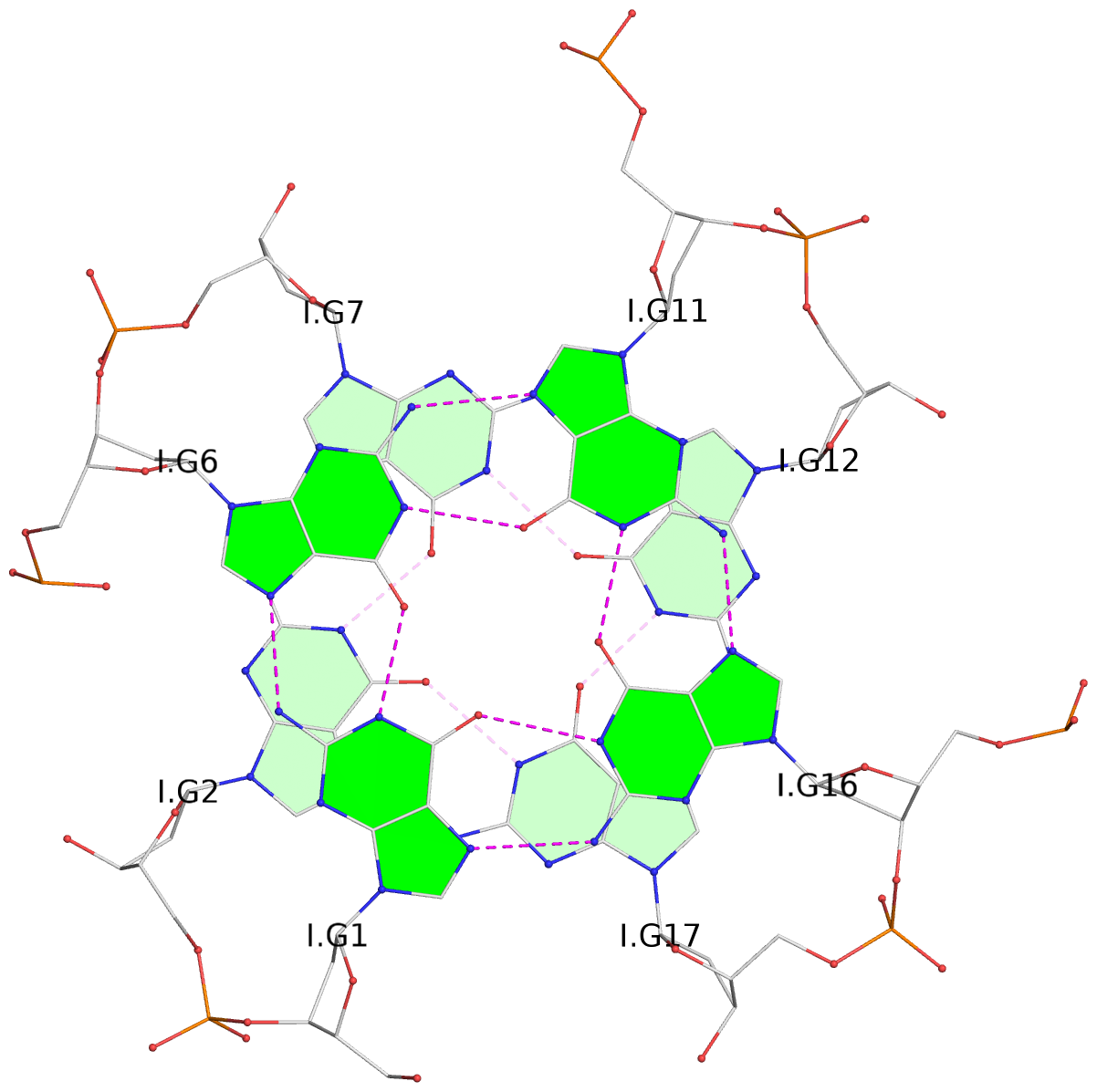
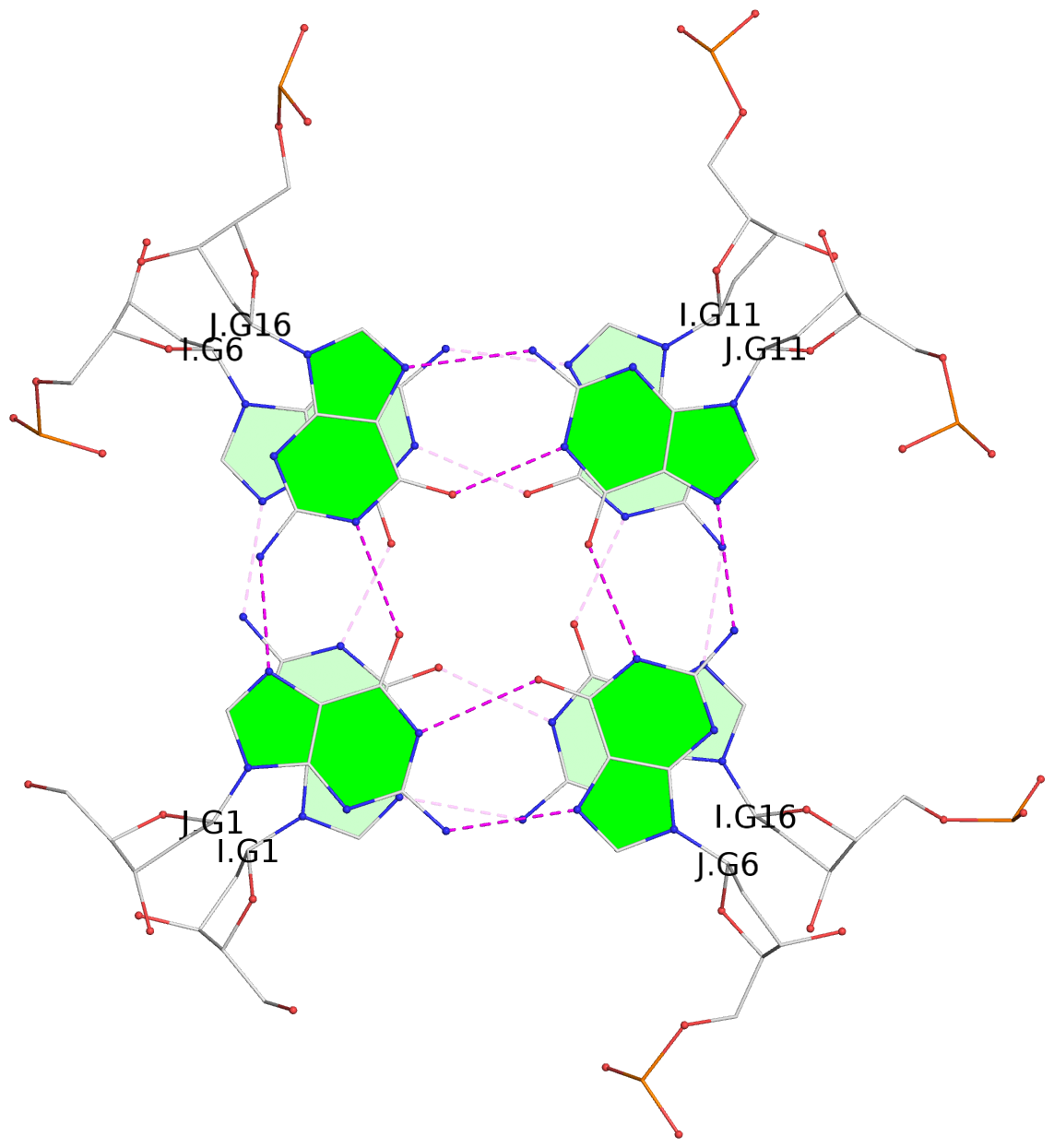
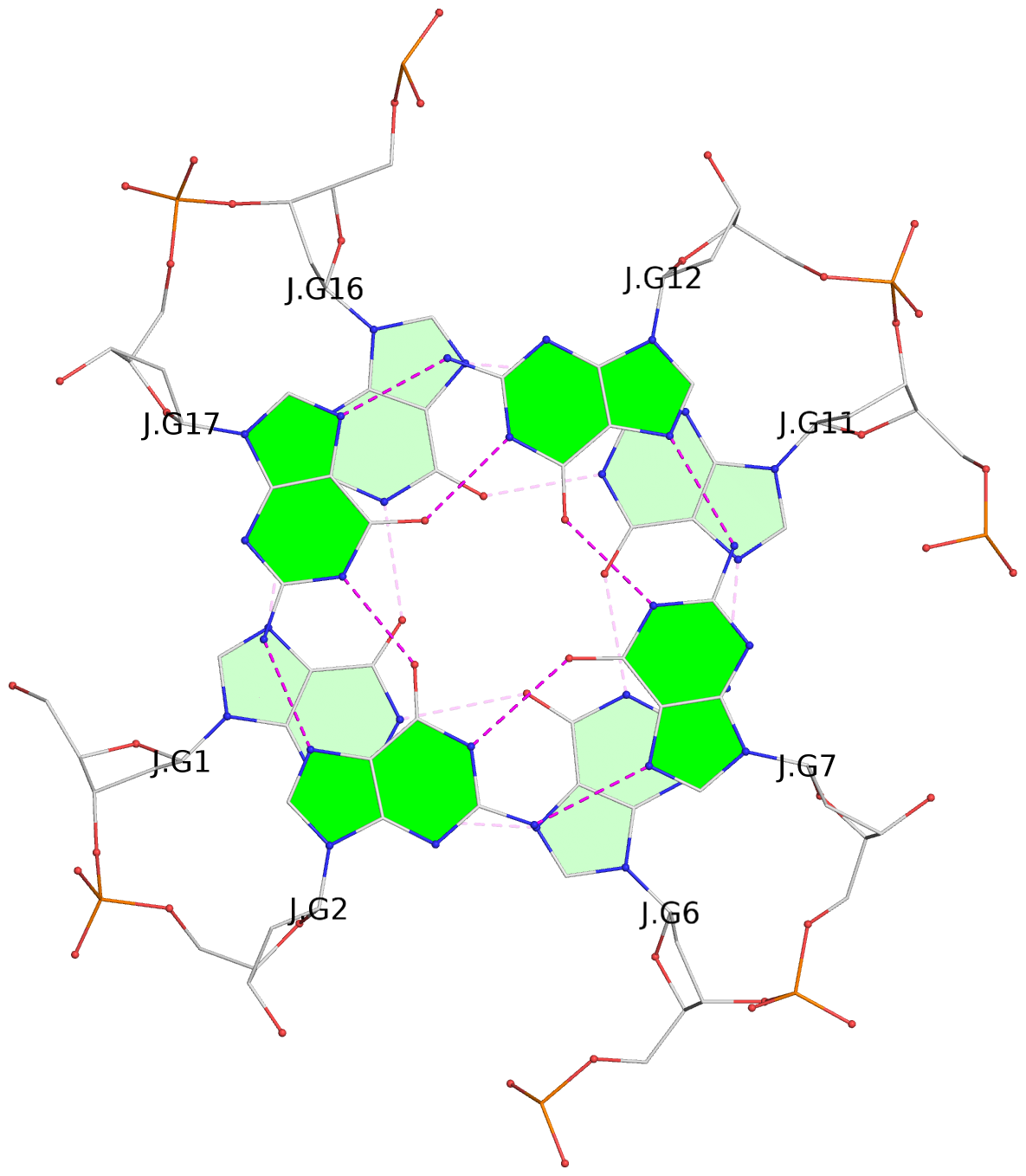
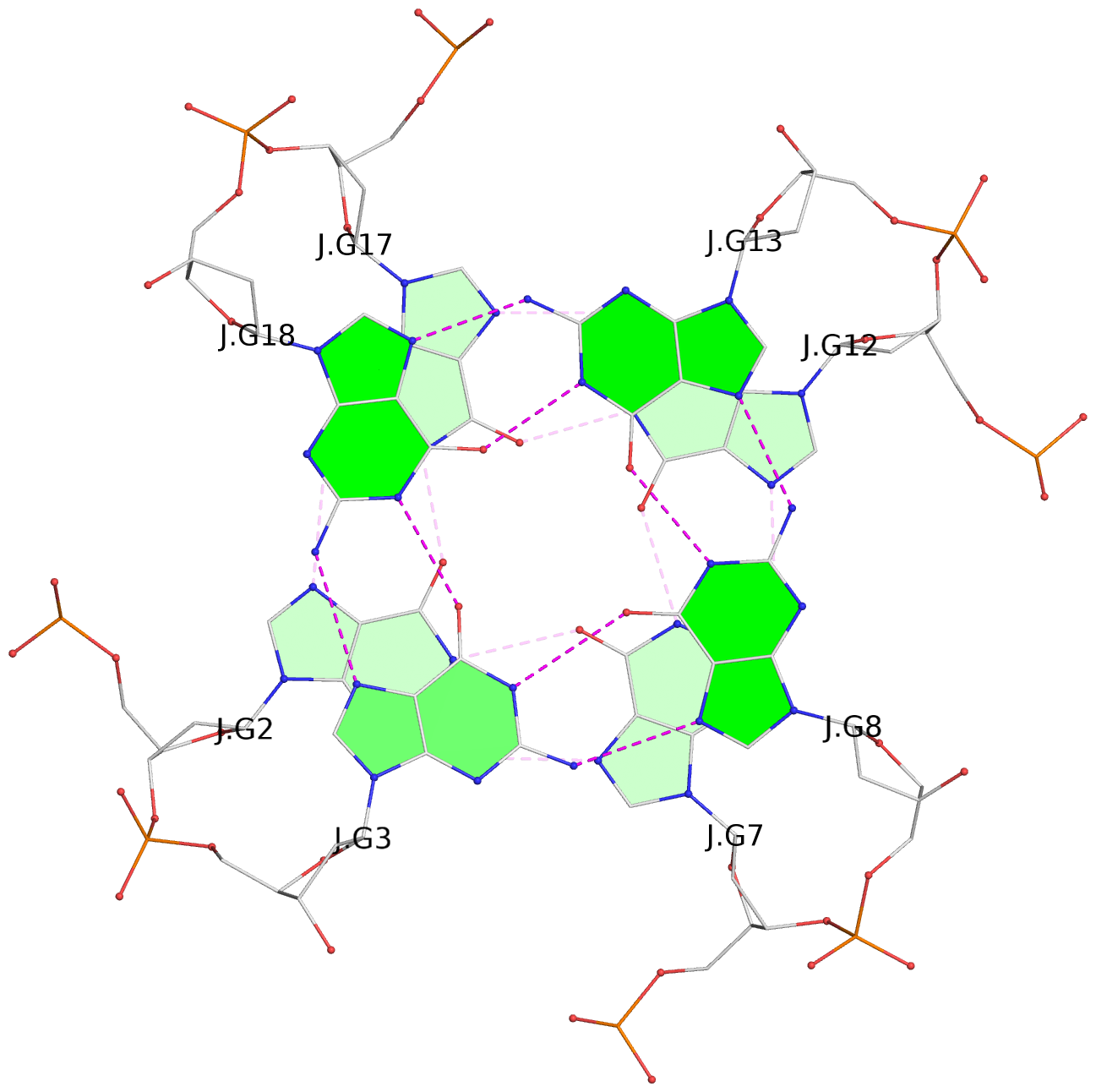
List of 30 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.071 type=planar nts=4 GGGG A.DG1,A.DG6,A.DG11,A.DG16 2 glyco-bond=---- sugar=---- groove=---- planarity=0.080 type=planar nts=4 GGGG A.DG2,A.DG7,A.DG12,A.DG17 3 glyco-bond=---- sugar=---- groove=---- planarity=0.274 type=bowl nts=4 GGGG A.DG3,A.DG8,A.DG13,A.DG18 4 glyco-bond=---- sugar=---- groove=---- planarity=0.085 type=planar nts=4 GGGG B.DG1,B.DG6,B.DG11,B.DG16 5 glyco-bond=---- sugar=---- groove=---- planarity=0.107 type=planar nts=4 GGGG B.DG2,B.DG7,B.DG12,B.DG17 6 glyco-bond=---- sugar=---- groove=---- planarity=0.269 type=bowl nts=4 GGGG B.DG3,B.DG8,B.DG13,B.DG18 7 glyco-bond=---- sugar=-.-- groove=---- planarity=0.129 type=planar nts=4 GGGG C.DG1,C.DG6,C.DG11,C.DG16 8 glyco-bond=---- sugar=---- groove=---- planarity=0.141 type=planar nts=4 GGGG C.DG2,C.DG7,C.DG12,C.DG17 9 glyco-bond=---- sugar=---- groove=---- planarity=0.236 type=bowl nts=4 GGGG C.DG3,C.DG8,C.DG13,C.DG18 10 glyco-bond=---- sugar=---- groove=---- planarity=0.112 type=planar nts=4 GGGG D.DG1,D.DG6,D.DG11,D.DG16 11 glyco-bond=---- sugar=---- groove=---- planarity=0.091 type=planar nts=4 GGGG D.DG2,D.DG7,D.DG12,D.DG17 12 glyco-bond=---- sugar=---- groove=---- planarity=0.362 type=bowl nts=4 GGGG D.DG3,D.DG8,D.DG13,D.DG18 13 glyco-bond=---- sugar=---- groove=---- planarity=0.058 type=planar nts=4 GGGG E.DG1,E.DG6,E.DG11,E.DG16 14 glyco-bond=---- sugar=---- groove=---- planarity=0.105 type=planar nts=4 GGGG E.DG2,E.DG7,E.DG12,E.DG17 15 glyco-bond=---- sugar=---- groove=---- planarity=0.209 type=other nts=4 GGGG E.DG3,E.DG8,E.DG13,E.DG18 16 glyco-bond=---- sugar=---- groove=---- planarity=0.083 type=planar nts=4 GGGG F.DG1,F.DG6,F.DG11,F.DG16 17 glyco-bond=---- sugar=---- groove=---- planarity=0.067 type=planar nts=4 GGGG F.DG2,F.DG7,F.DG12,F.DG17 18 glyco-bond=---- sugar=---- groove=---- planarity=0.298 type=bowl nts=4 GGGG F.DG3,F.DG8,F.DG13,F.DG18 19 glyco-bond=---- sugar=---- groove=---- planarity=0.099 type=planar nts=4 GGGG G.DG1,G.DG6,G.DG11,G.DG16 20 glyco-bond=---- sugar=---- groove=---- planarity=0.139 type=planar nts=4 GGGG G.DG2,G.DG7,G.DG12,G.DG17 21 glyco-bond=---- sugar=---- groove=---- planarity=0.294 type=bowl nts=4 GGGG G.DG3,G.DG8,G.DG13,G.DG18 22 glyco-bond=---- sugar=---. groove=---- planarity=0.070 type=planar nts=4 GGGG H.DG1,H.DG6,H.DG11,H.DG16 23 glyco-bond=---- sugar=---- groove=---- planarity=0.074 type=planar nts=4 GGGG H.DG2,H.DG7,H.DG12,H.DG17 24 glyco-bond=---- sugar=---- groove=---- planarity=0.264 type=bowl nts=4 GGGG H.DG3,H.DG8,H.DG13,H.DG18 25 glyco-bond=---- sugar=---- groove=---- planarity=0.089 type=planar nts=4 GGGG I.DG1,I.DG6,I.DG11,I.DG16 26 glyco-bond=---- sugar=---- groove=---- planarity=0.167 type=other nts=4 GGGG I.DG2,I.DG7,I.DG12,I.DG17 27 glyco-bond=---- sugar=---- groove=---- planarity=0.215 type=other nts=4 GGGG I.DG3,I.DG8,I.DG13,I.DG18 28 glyco-bond=---- sugar=---- groove=---- planarity=0.042 type=planar nts=4 GGGG J.DG1,J.DG6,J.DG11,J.DG16 29 glyco-bond=---- sugar=---- groove=---- planarity=0.071 type=planar nts=4 GGGG J.DG2,J.DG7,J.DG12,J.DG17 30 glyco-bond=---- sugar=---- groove=---- planarity=0.324 type=bowl nts=4 GGGG J.DG3,J.DG8,J.DG13,J.DG18
List of 5 G4-helices
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
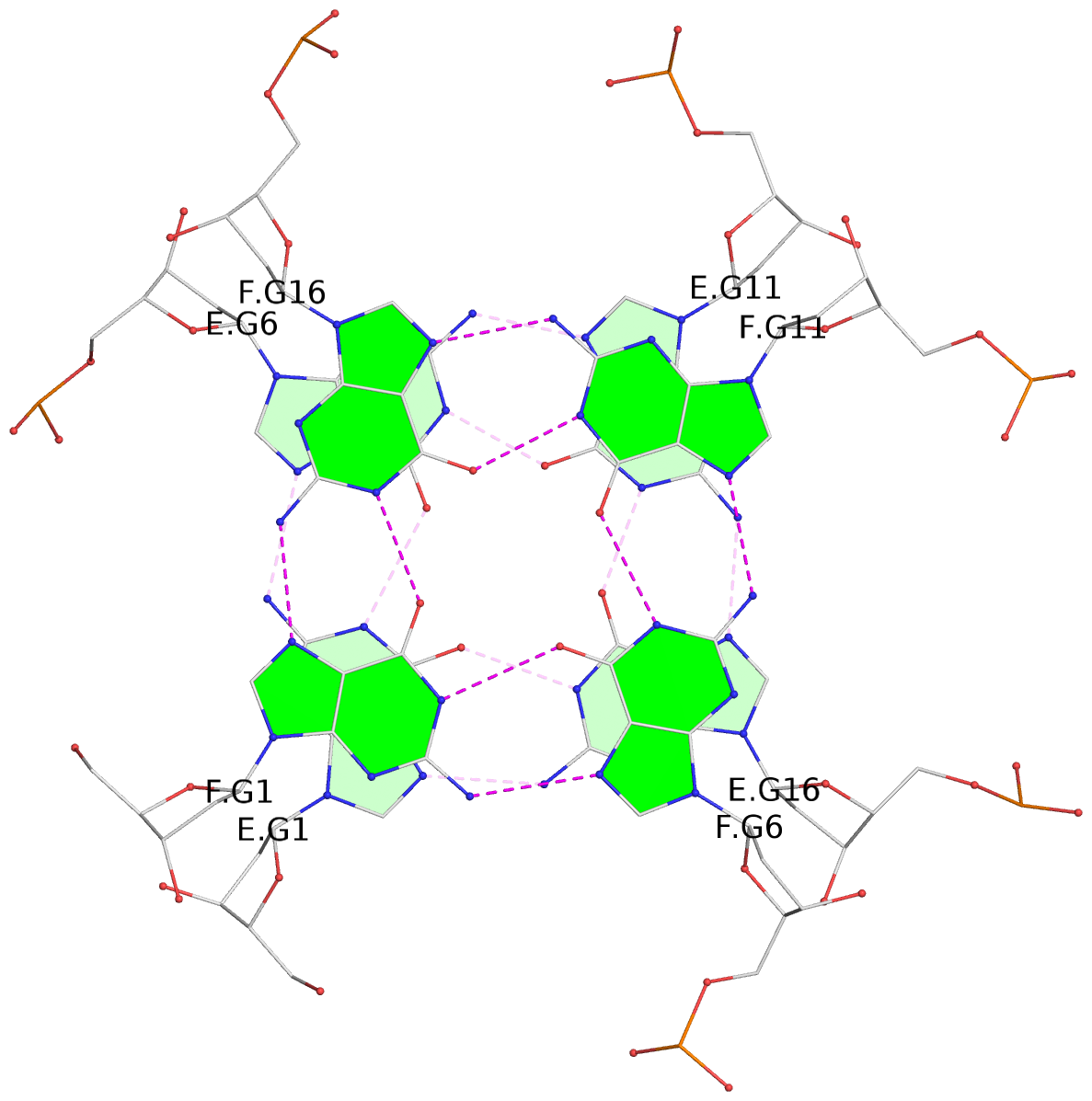
Helix#1, 6 G-tetrad layers, inter-molecular, with 2 stems
Helix#2, 6 G-tetrad layers, inter-molecular, with 2 stems
Helix#3, 6 G-tetrad layers, inter-molecular, with 2 stems
Helix#4, 6 G-tetrad layers, inter-molecular, with 2 stems
Helix#5, 6 G-tetrad layers, inter-molecular, with 2 stems
List of 10 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 3 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 3(-P-P-P), parallel(4+0)
Stem#2, 3 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 3(-P-P-P), parallel(4+0)
Stem#3, 3 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 3(-P-P-P), parallel(4+0)
Stem#4, 3 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 3(-P-P-P), parallel(4+0)
Stem#5, 3 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 3(-P-P-P), parallel(4+0)
Stem#6, 3 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 3(-P-P-P), parallel(4+0)
Stem#7, 3 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 3(-P-P-P), parallel(4+0)
Stem#8, 3 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 3(-P-P-P), parallel(4+0)
Stem#9, 3 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 3(-P-P-P), parallel(4+0)
Stem#10, 3 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 3(-P-P-P), parallel(4+0)
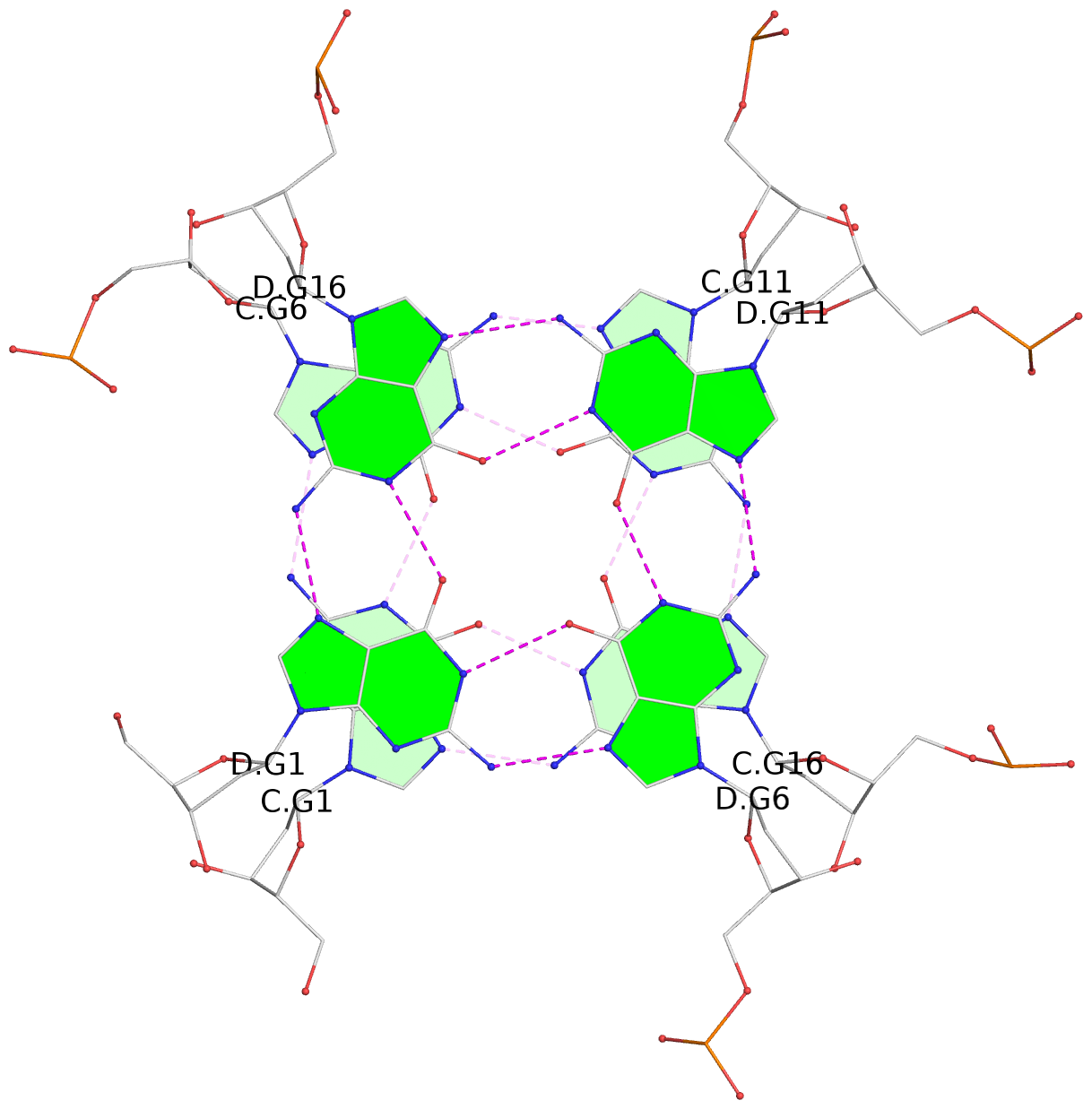
List of 5 G4 coaxial stacks
1 G4 helix#1 contains 2 G4 stems: [#1,#2] [5'/5'] 2 G4 helix#2 contains 2 G4 stems: [#3,#4] [5'/5'] 3 G4 helix#3 contains 2 G4 stems: [#5,#6] [5'/5'] 4 G4 helix#4 contains 2 G4 stems: [#7,#8] [5'/5'] 5 G4 helix#5 contains 2 G4 stems: [#9,#10] [5'/5']