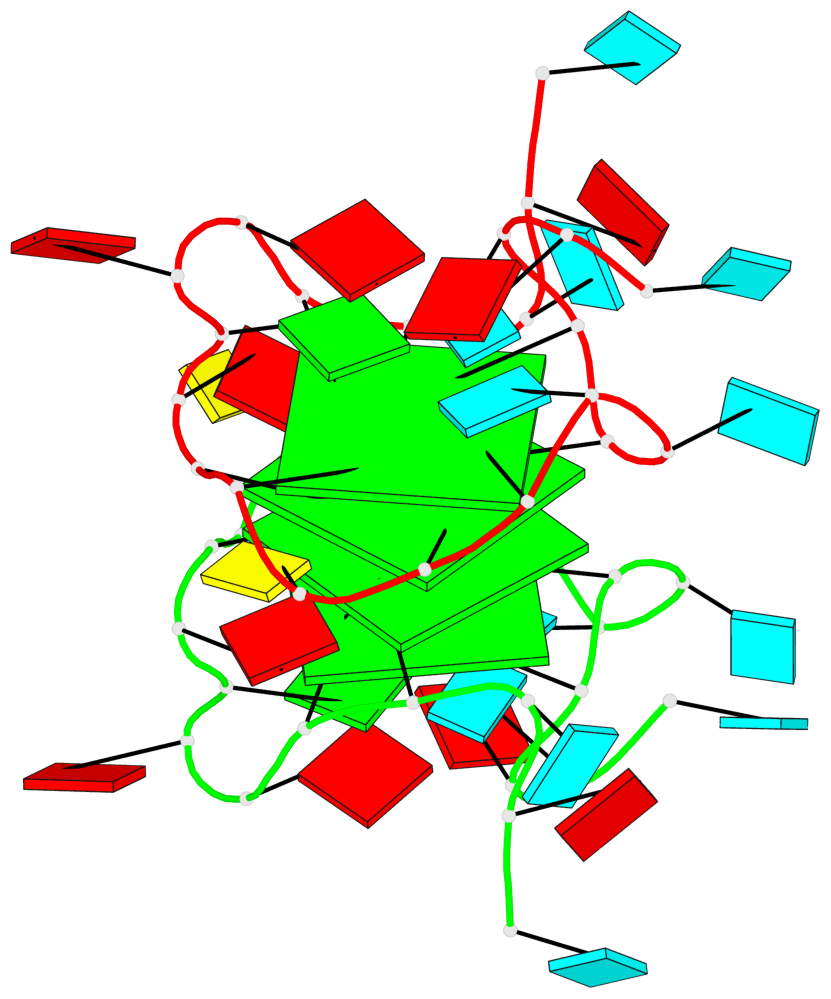
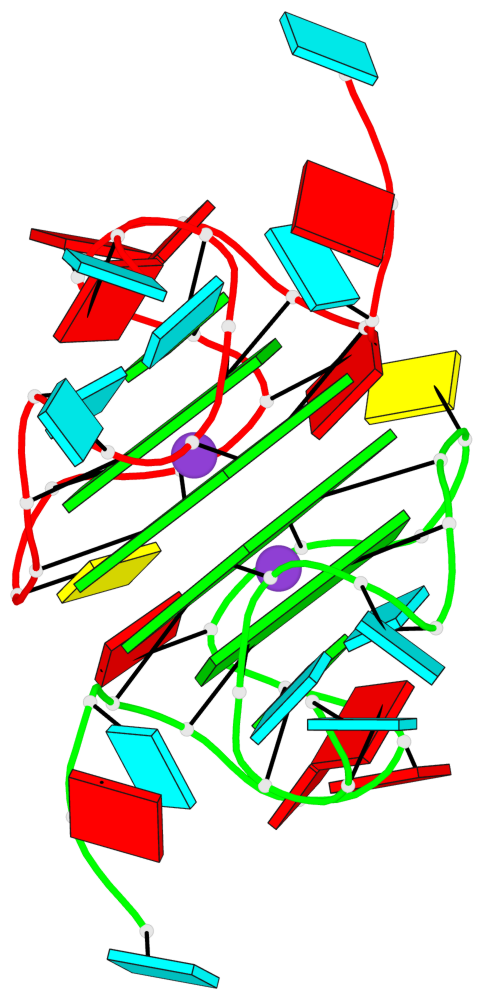
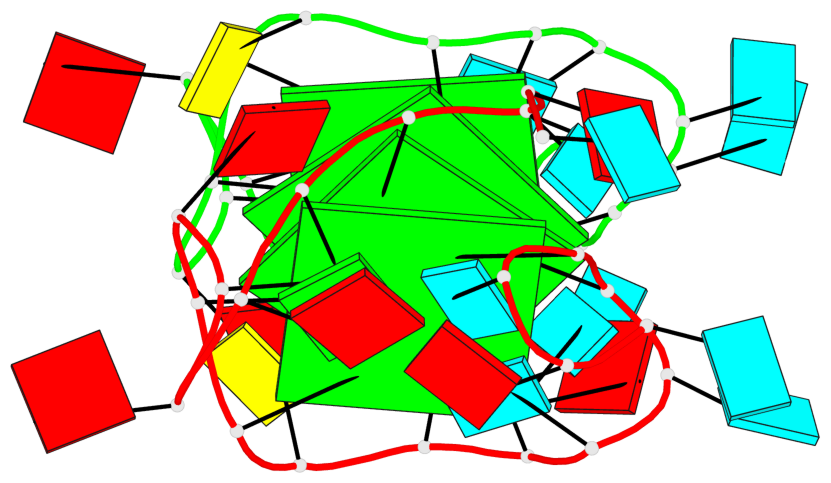
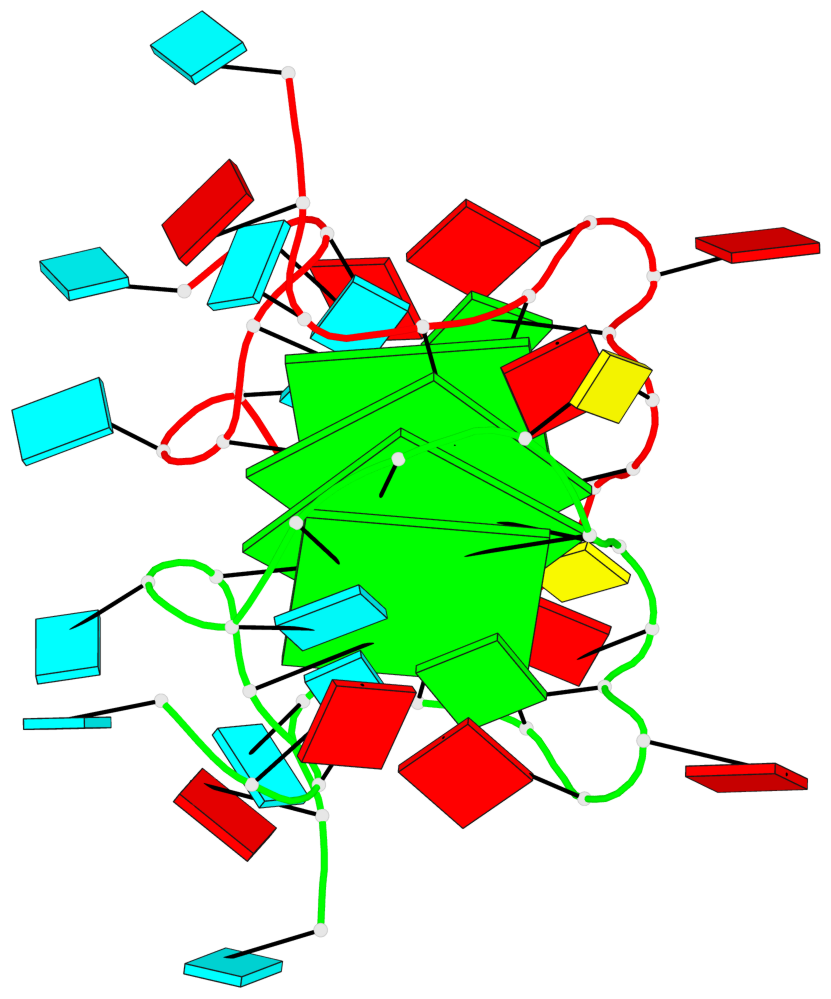
Detailed DSSR results for the G-quadruplex: PDB entry 8utg
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 8utg
- Class
- RNA
- Method
- X-ray (1.97 Å)
- Summary
- An RNA g-quadruplex from the ns5 gene in the west nile virus genome
- Reference
- Terrell JR, Le TT, Paul A, Brinton MA, Wilson WD, Poon GMK, Germann MW, Siemer JL (2024): "Structure of an RNA G-quadruplex from the West Nile virus genome." Nat Commun, 15, 5428. doi: 10.1038/s41467-024-49761-5.
- Abstract
- Potential G-quadruplex sites have been identified in the genomes of DNA and RNA viruses and proposed as regulatory elements. The genus Orthoflavivirus contains arthropod-transmitted, positive-sense, single-stranded RNA viruses that cause significant human disease globally. Computational studies have identified multiple potential G-quadruplex sites that are conserved across members of this genus. Subsequent biophysical studies established that some G-quadruplexes predicted in Zika and tickborne encephalitis virus genomes can form and known quadruplex binders reduced viral yields from cells infected with these viruses. The susceptibility of RNA to degradation and the variability of loop regions have made structure determination challenging. Despite these difficulties, we report a high-resolution structure of the NS5-B quadruplex from the West Nile virus genome. Analysis reveals two stacked tetrads that are further stabilized by a stacked triad and transient noncanonical base pairing. This structure expands the landscape of solved RNA quadruplex structures and demonstrates the diversity and complexity of biological quadruplexes. We anticipate that the availability of this structure will assist in solving further viral RNA quadruplexes and provides a model for a conserved antiviral target in Orthoflavivirus genomes.
- G4 notes
- 4 G-tetrads, 1 G4 helix, 2 G4 stems, 1 G4 coaxial stack, 2(-P-P-P), parallel(4+0), UUUU, coaxial interfaces: 5'/5'
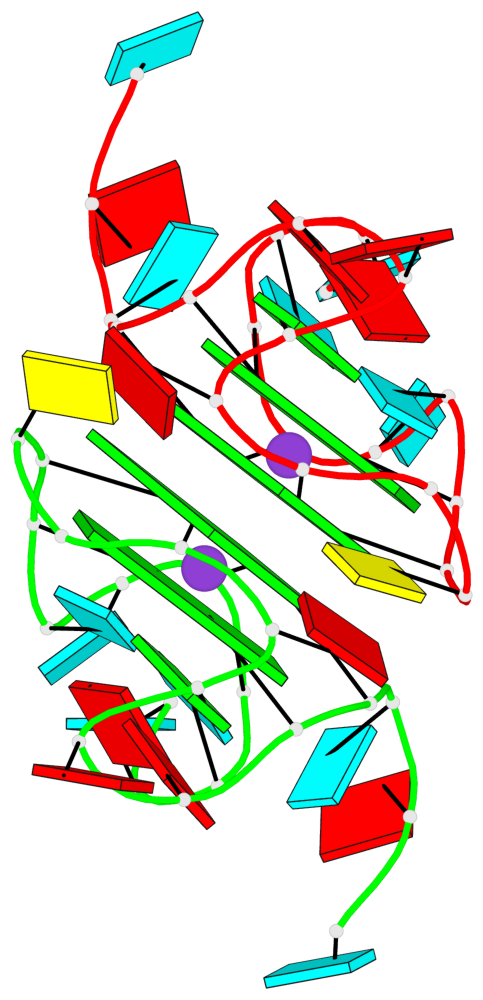
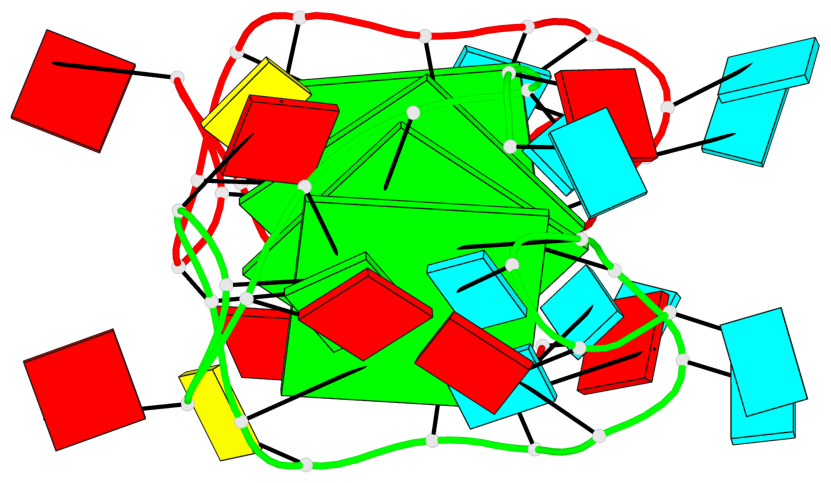
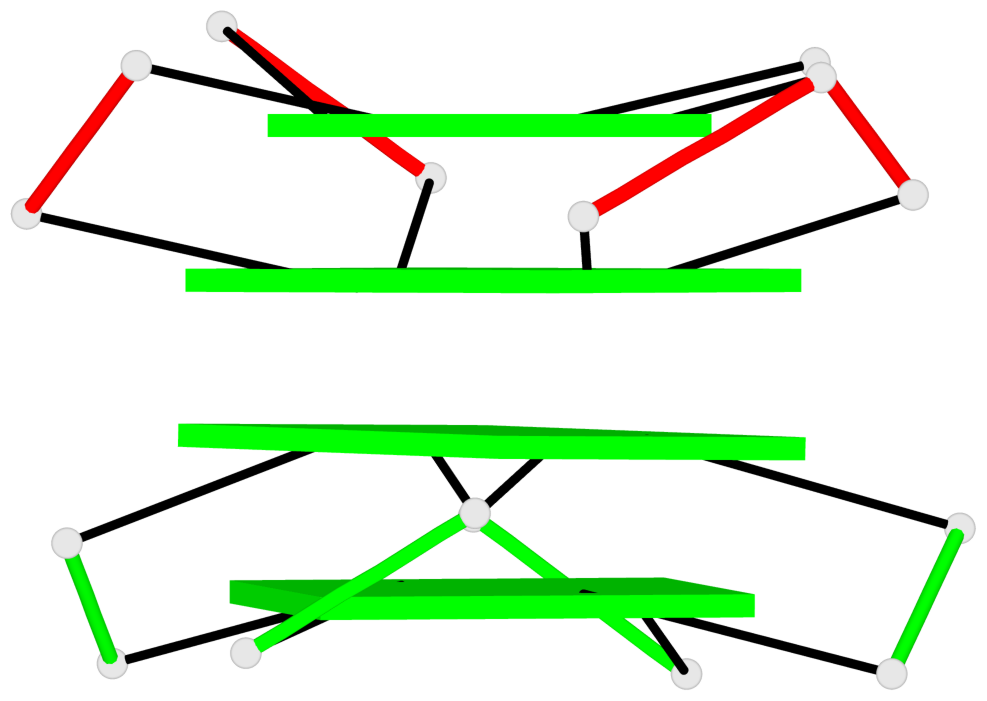
Base-block schematics in six views
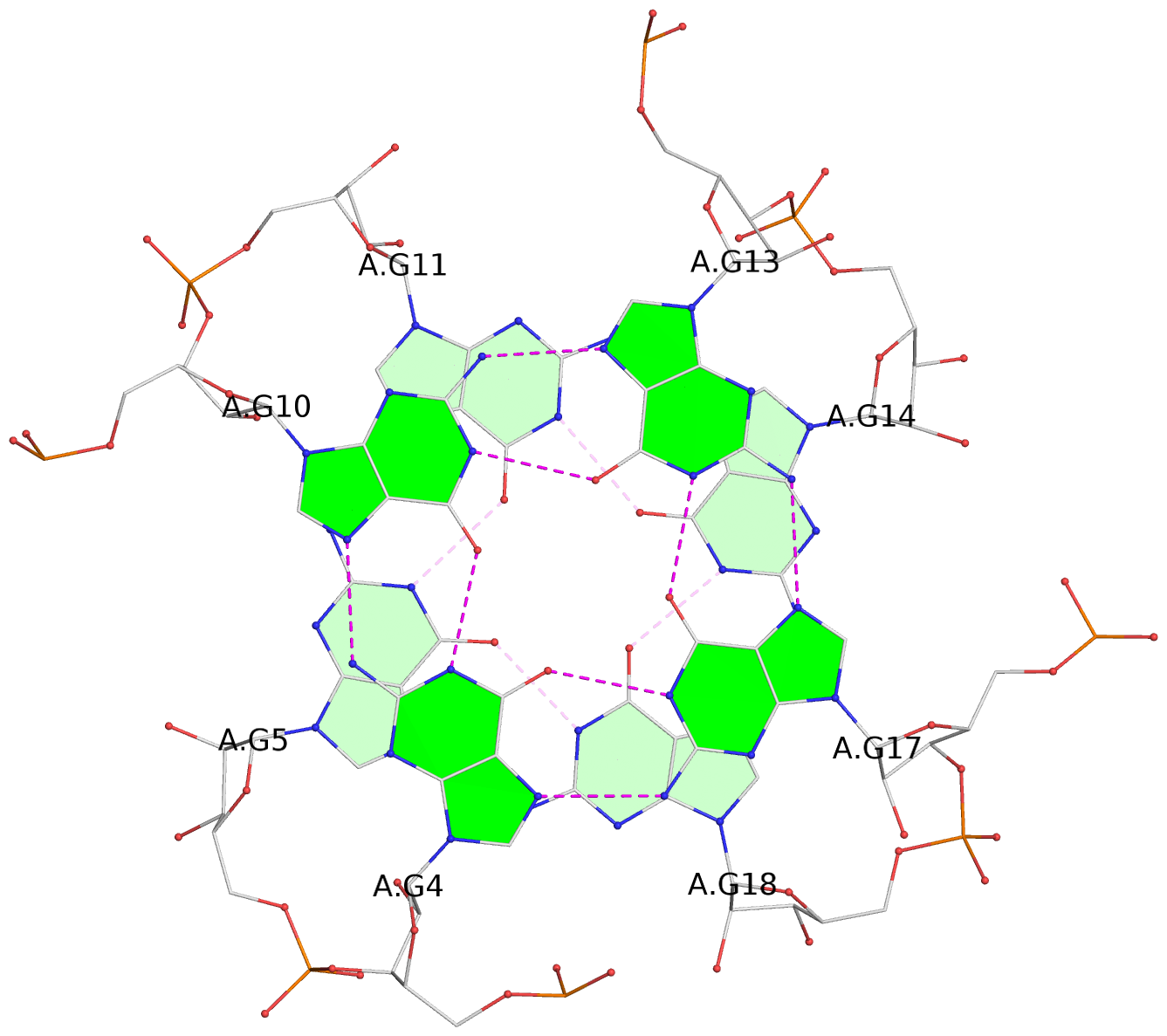
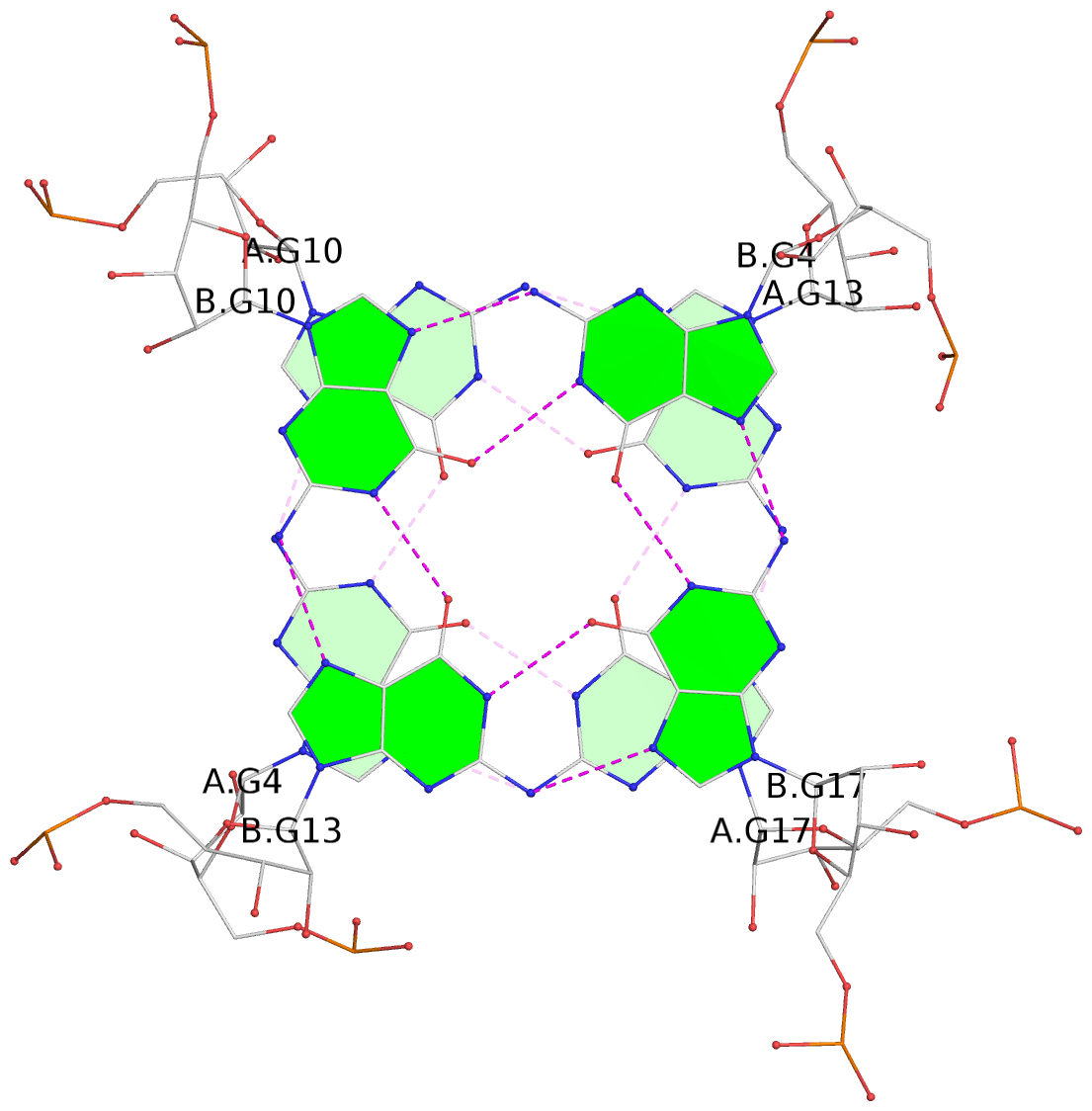
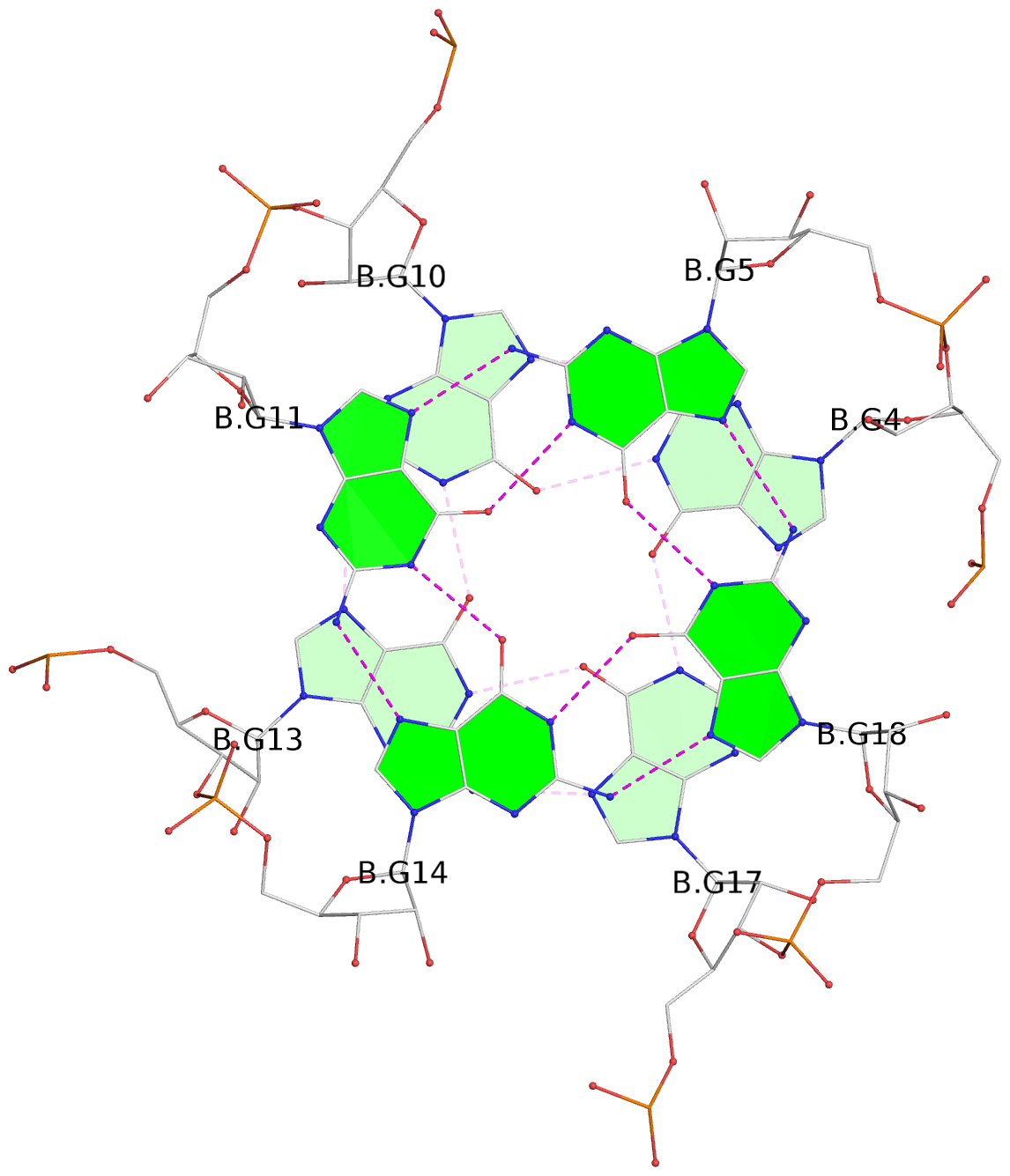
List of 4 G-tetrads
1 glyco-bond=---- sugar=--33 groove=---- planarity=0.136 type=planar nts=4 GGGG A.G4,A.G10,A.G13,A.G17 2 glyco-bond=---- sugar=3-33 groove=---- planarity=0.168 type=other nts=4 GGGG A.G5,A.G11,A.G14,A.G18 3 glyco-bond=---- sugar=-333 groove=---- planarity=0.171 type=other nts=4 GGGG B.G4,B.G10,B.G13,B.G17 4 glyco-bond=---- sugar=3-33 groove=---- planarity=0.160 type=planar nts=4 GGGG B.G5,B.G11,B.G14,B.G18
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 4 G-tetrad layers, inter-molecular, with 2 stems
List of 2 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(-P-P-P), parallel(4+0)
Stem#2, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(-P-P-P), parallel(4+0)
List of 1 G4 coaxial stack
1 G4 helix#1 contains 2 G4 stems: [#1,#2] [5'/5']