Detailed DSSR results for the G-quadruplex: PDB entry 8x0s
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 8x0s
- Class
- RNA
- Method
- X-ray (2.956 Å)
- Summary
- Crystal structure of r(g4c2)2
- Reference
- Geng Y, Liu C, Xu N, Suen MC, Miao H, Xie Y, Zhang B, Chen X, Song Y, Wang Z, Cai Q, Zhu G (2024): "Crystal structure of a tetrameric RNA G-quadruplex formed by hexanucleotide repeat expansions of C9orf72 in ALS/FTD." Nucleic Acids Res., 52, 7961-7970. doi: 10.1093/nar/gkae473.
- Abstract
- The abnormal GGGGCC hexanucleotide repeat expansions (HREs) in C9orf72 cause the fatal neurodegenerative diseases including amyotrophic lateral sclerosis and frontotemporal dementia. The transcribed RNA HREs, short for r(G4C2)n, can form toxic RNA foci which sequestrate RNA binding proteins and impair RNA processing, ultimately leading to neurodegeneration. Here, we determined the crystal structure of r(G4C2)2, which folds into a parallel tetrameric G-quadruplex composed of two four-layer dimeric G-quadruplex via 5'-to-5' stacking in coordination with a K+ ion. Notably, the two C bases locate at 3'- end stack on the outer G-tetrad with the assistance of two additional K+ ions. The high-resolution structure reported here lays a foundation in understanding the mechanism of neurological toxicity of RNA HREs. Furthermore, the atomic details provide a structural basis for the development of potential therapeutic agents against the fatal neurodegenerative diseases ALS/FTD.
- G4 notes
- 8 G-tetrads, 1 G4 helix, 2 G4 stems, 1 G4 coaxial stack, parallel(4+0), UUUU, coaxial interfaces: 5'/5'
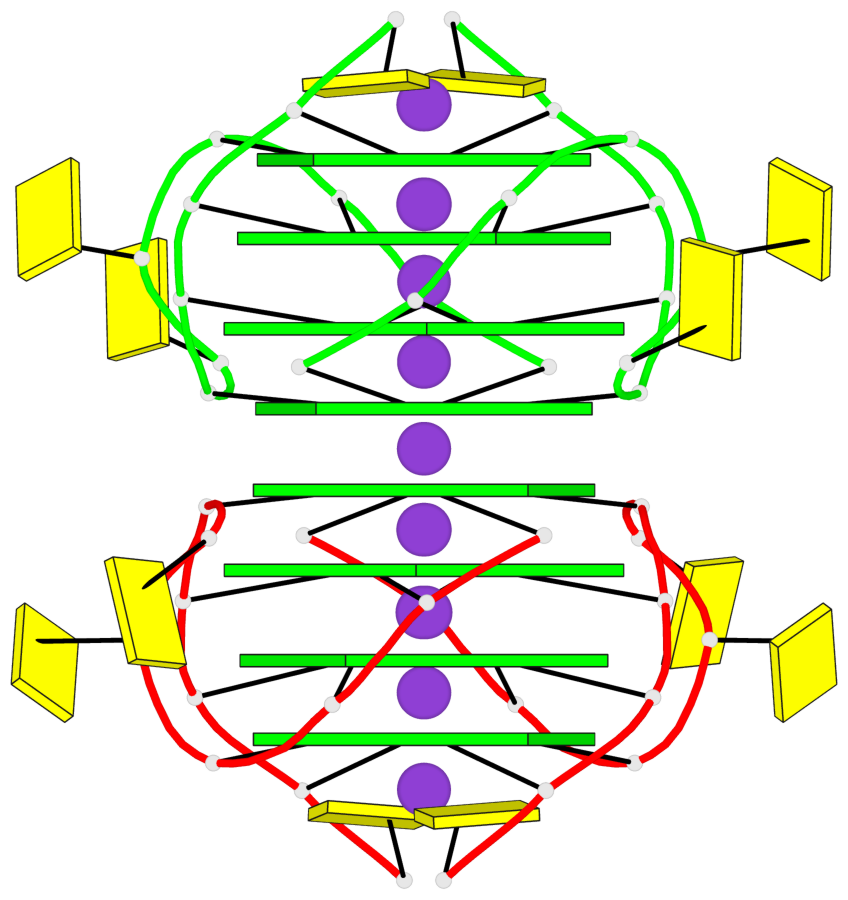
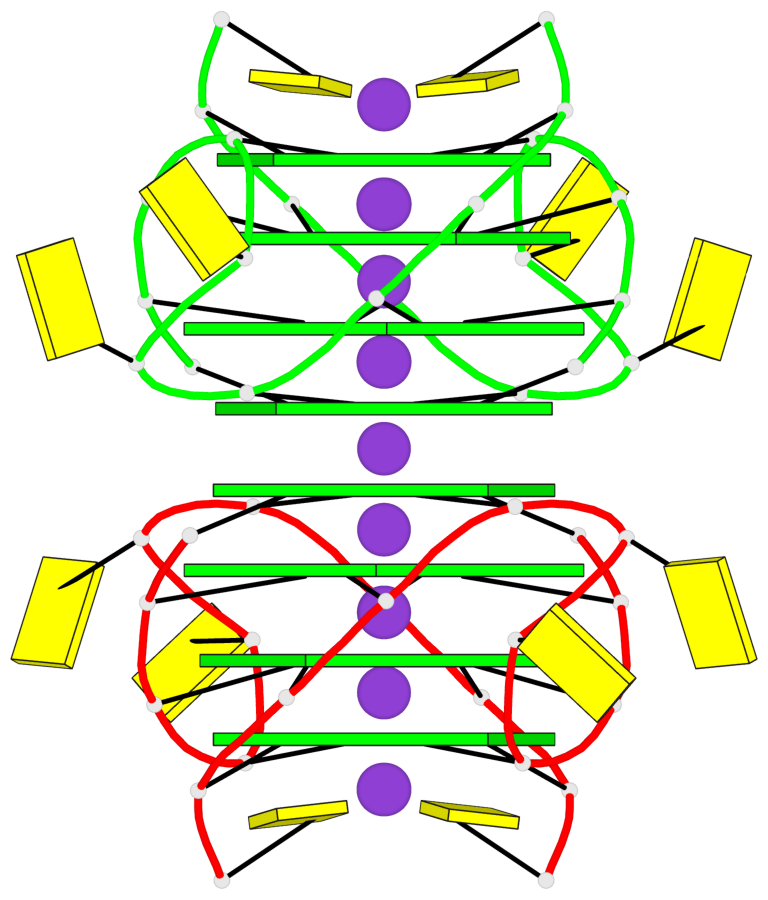
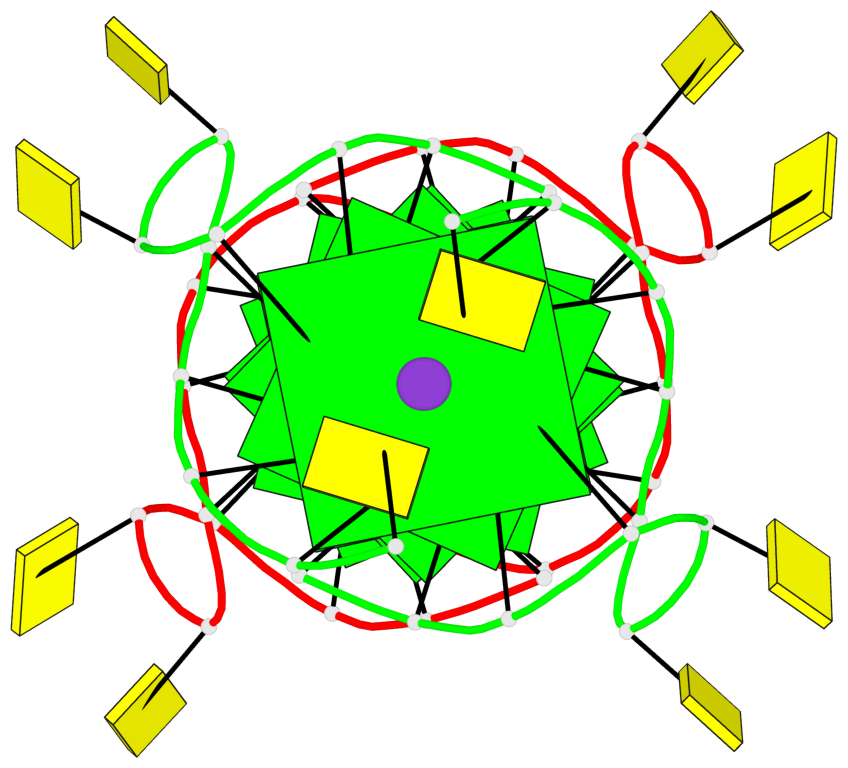
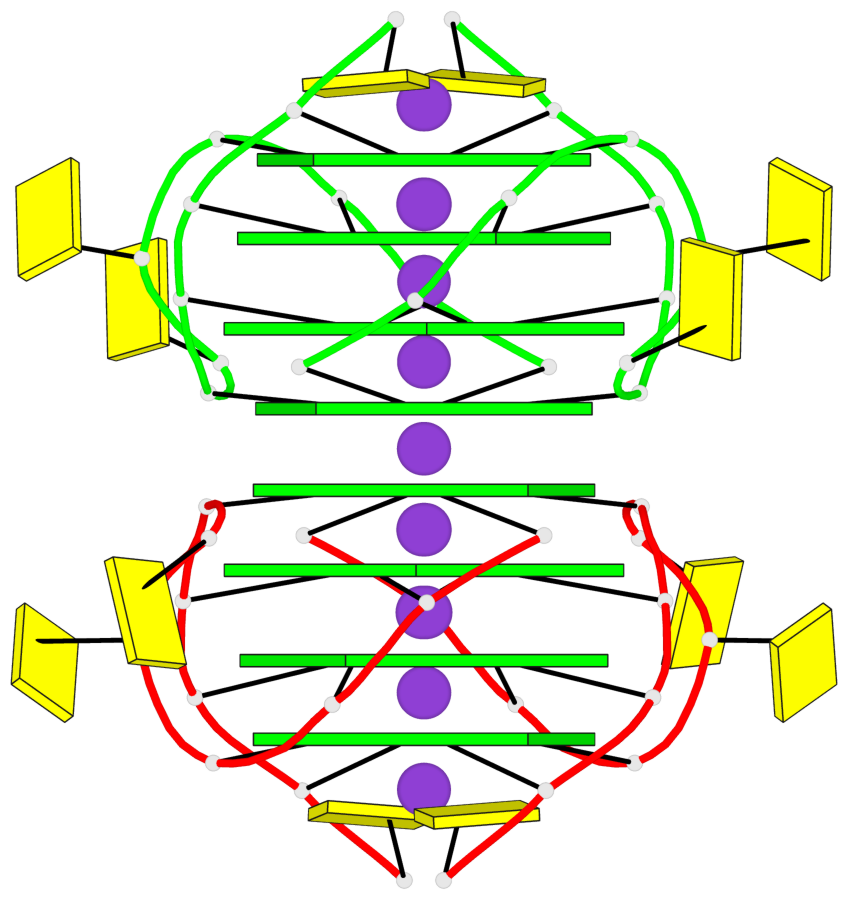
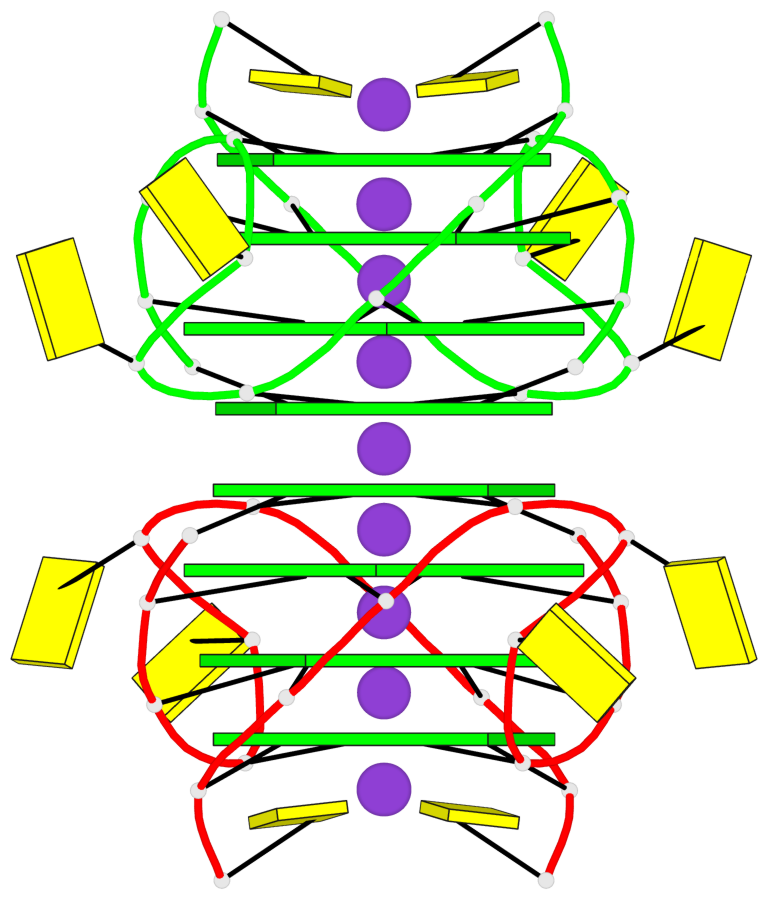
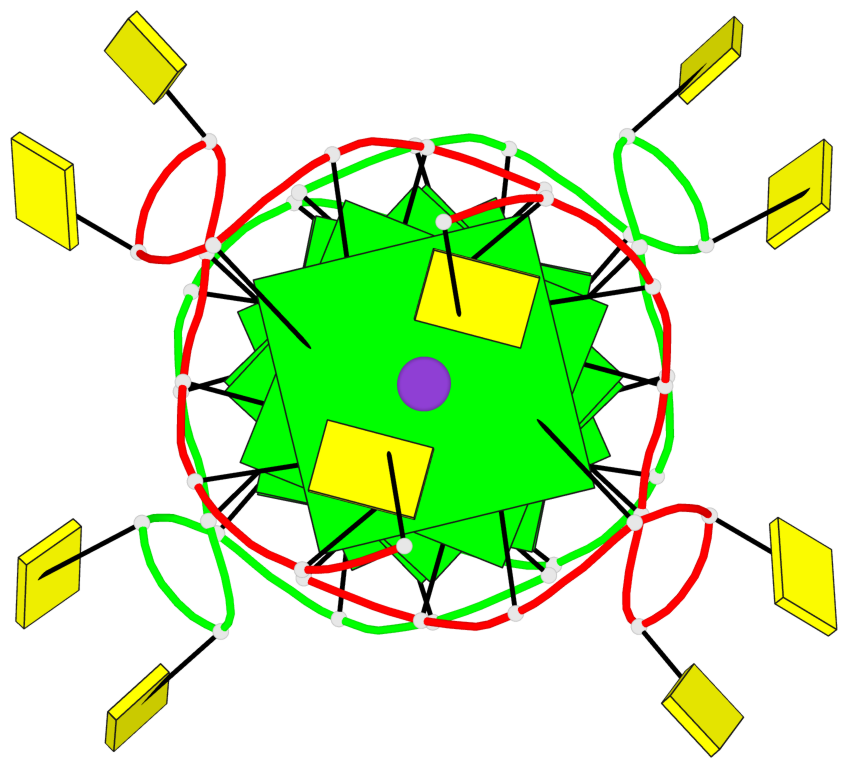
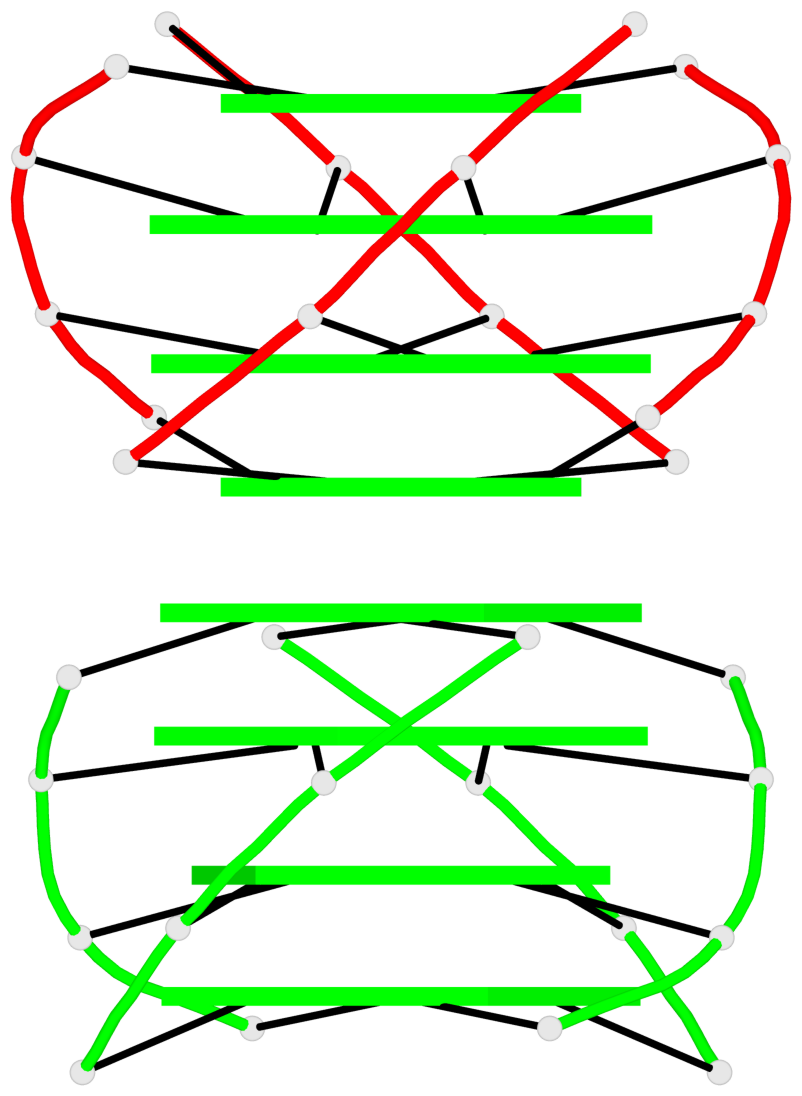
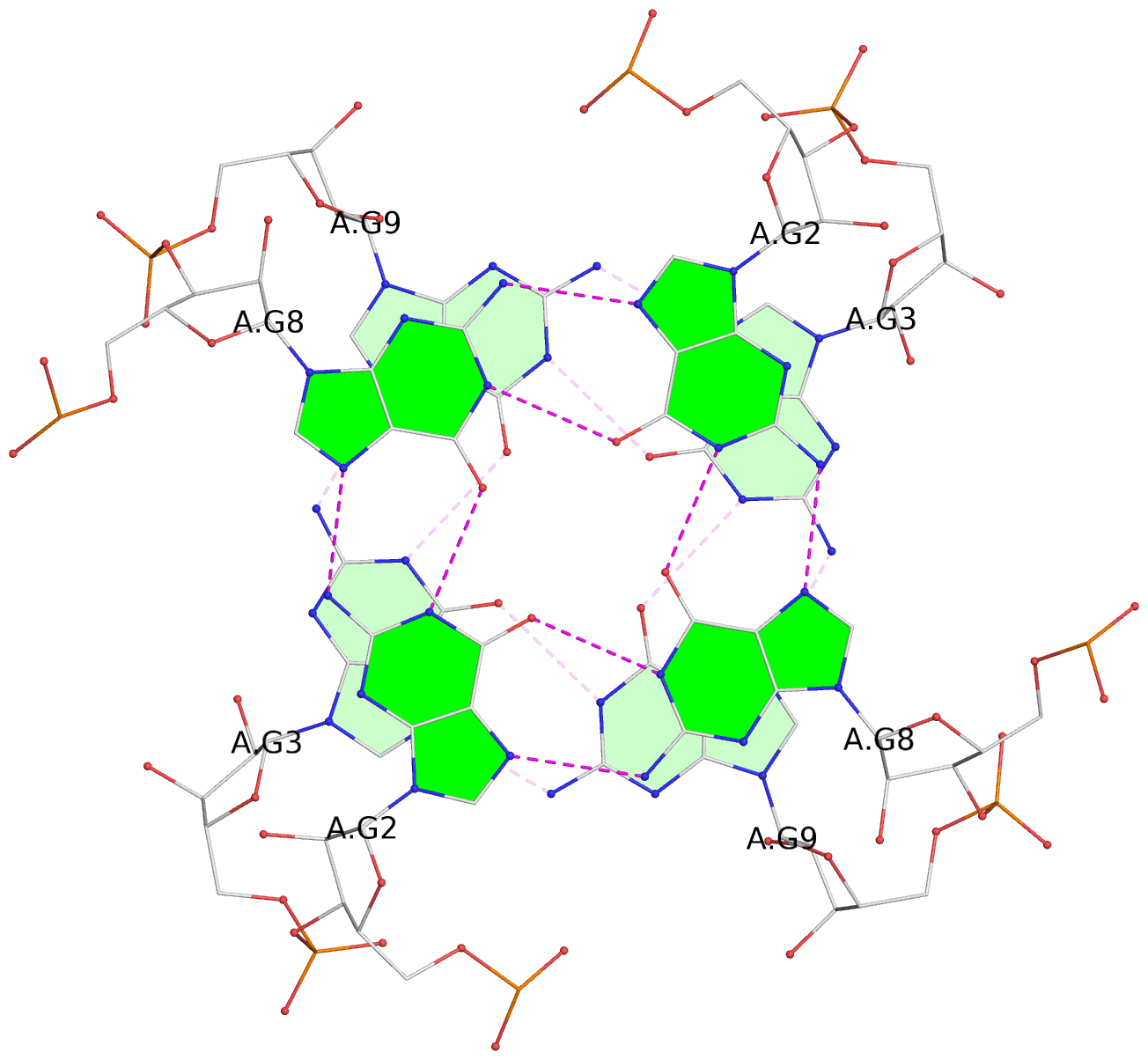
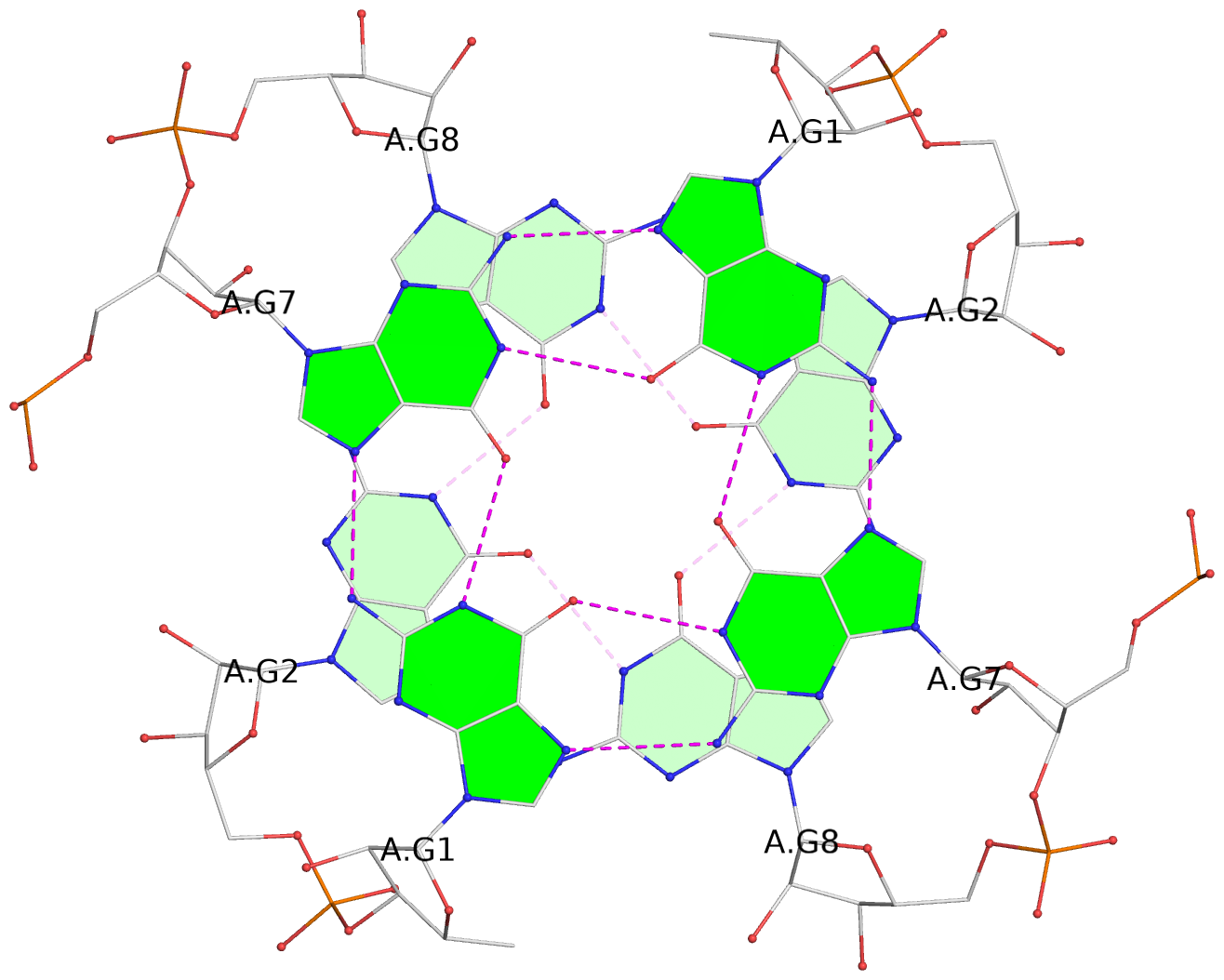
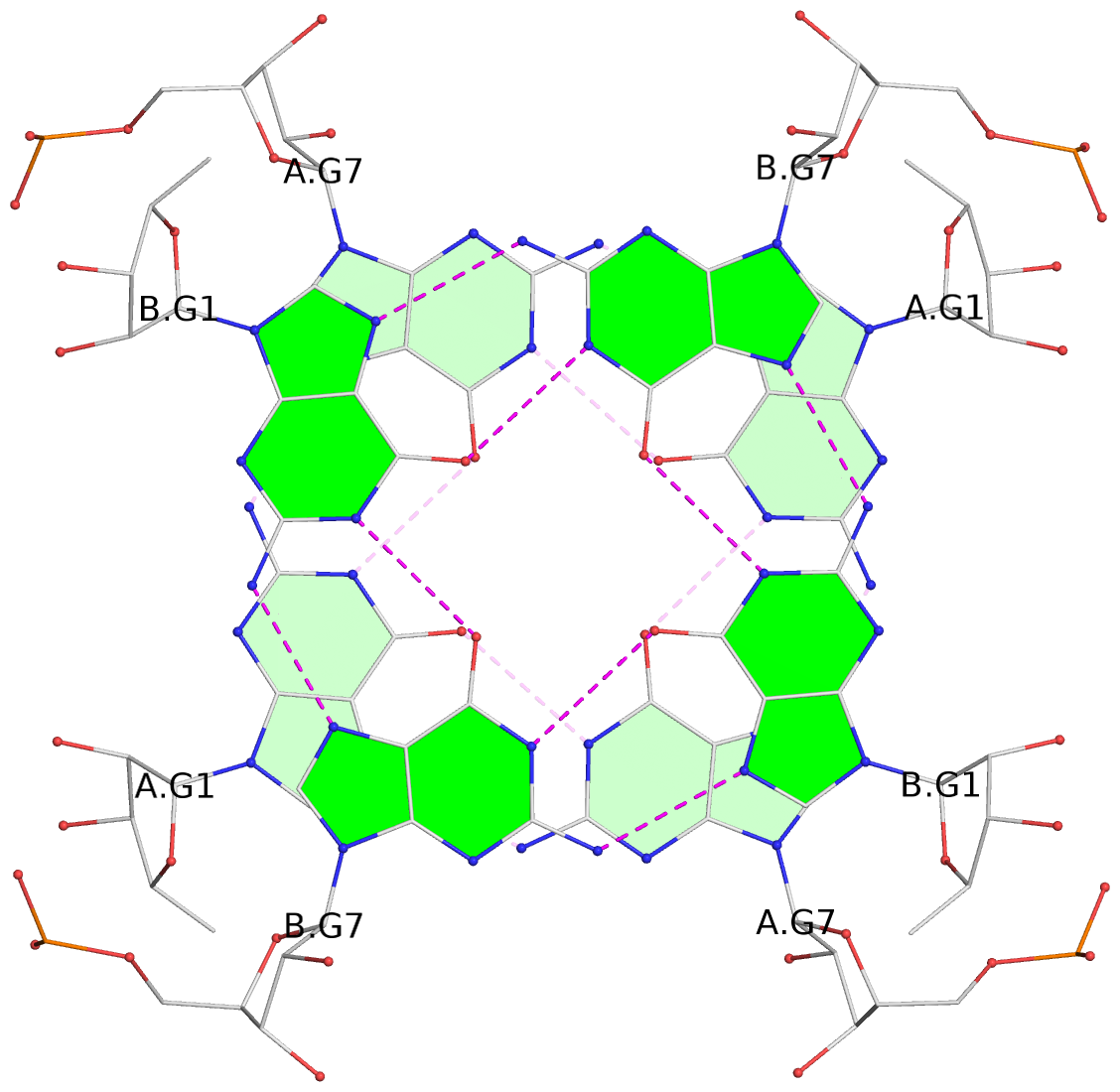
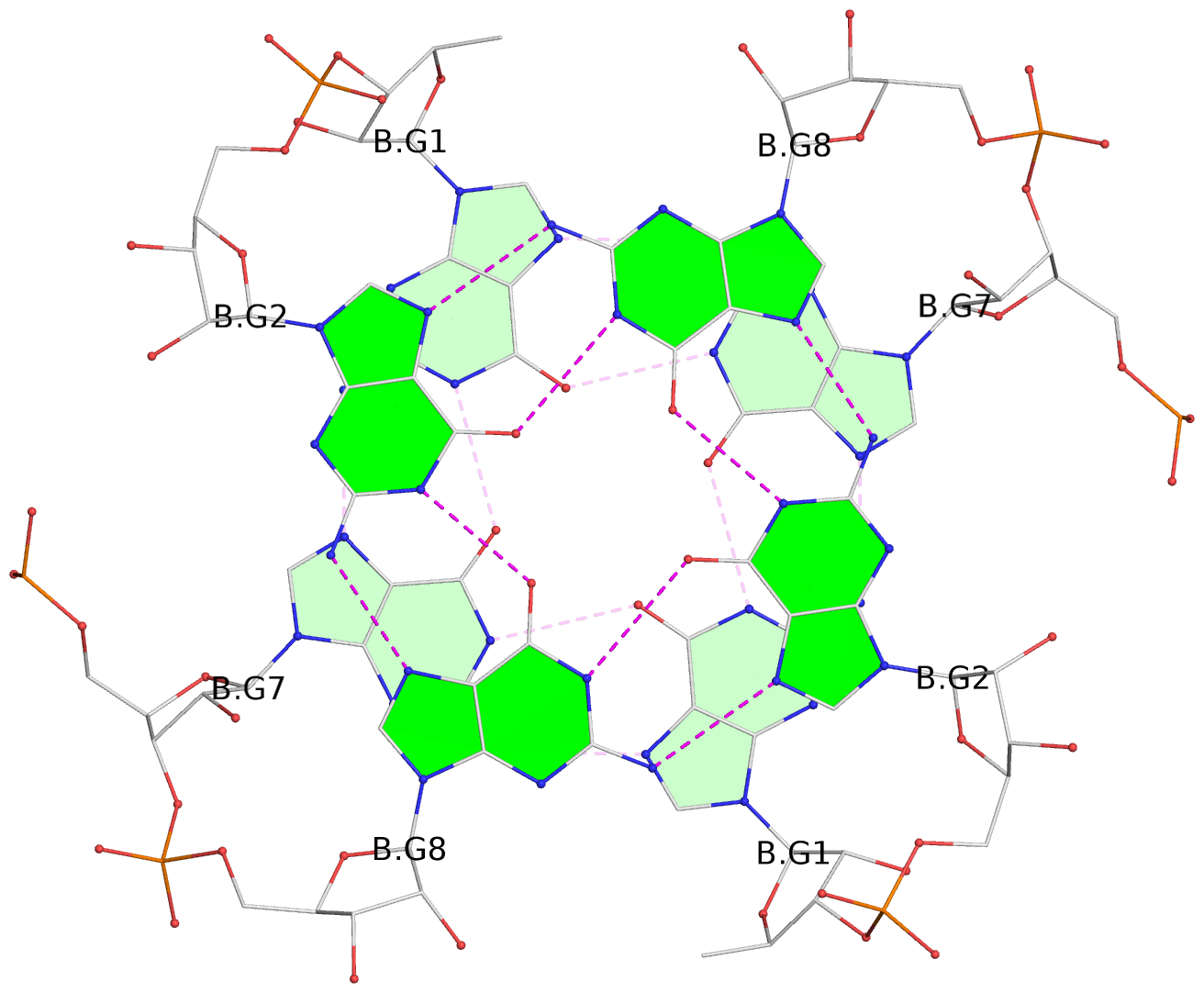
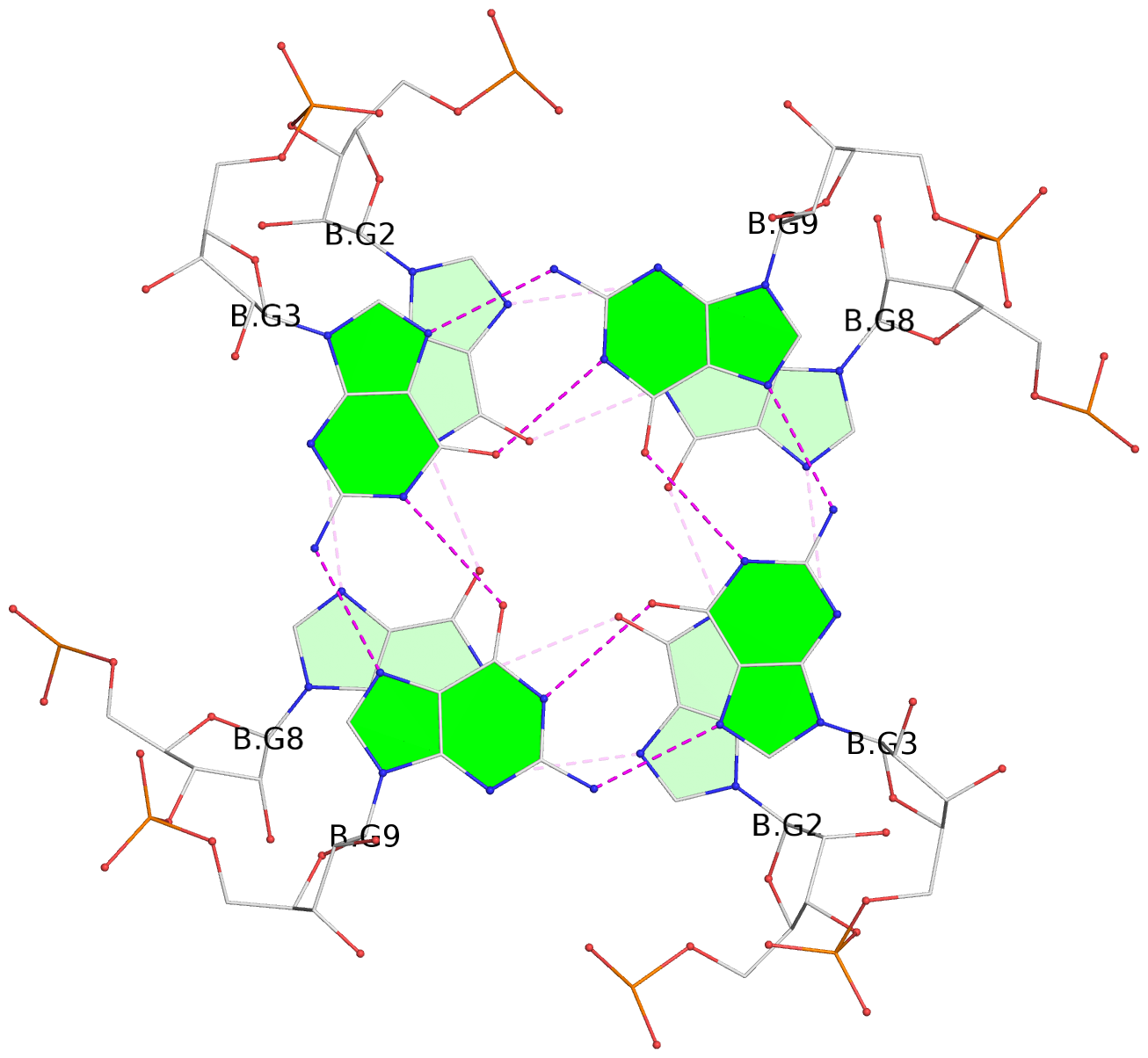
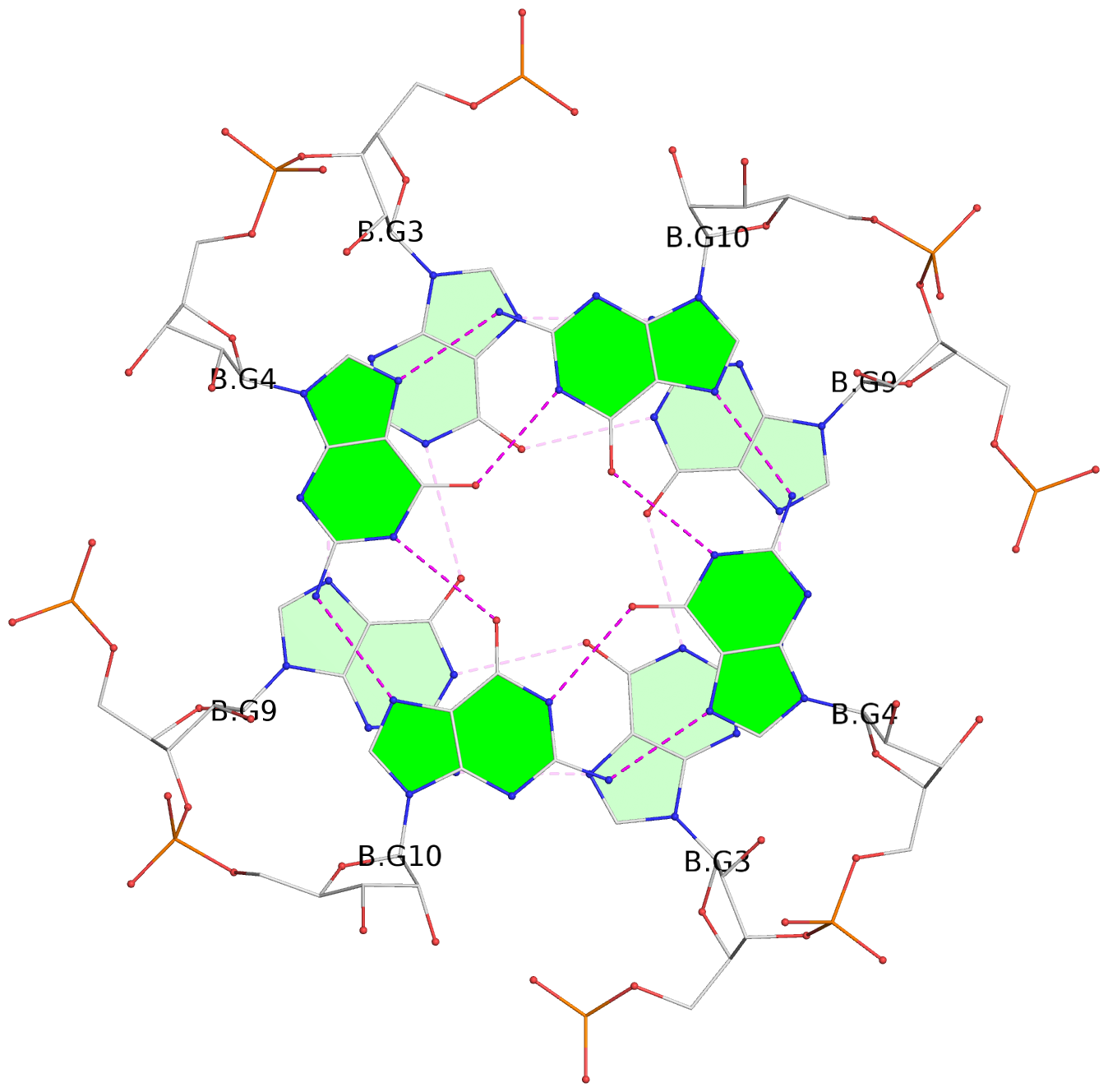
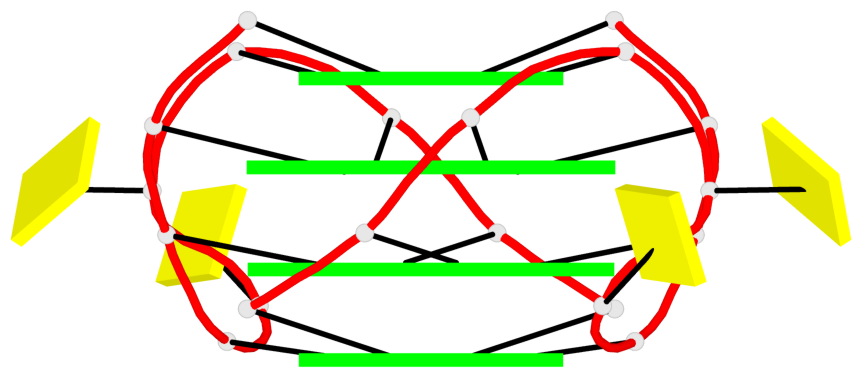
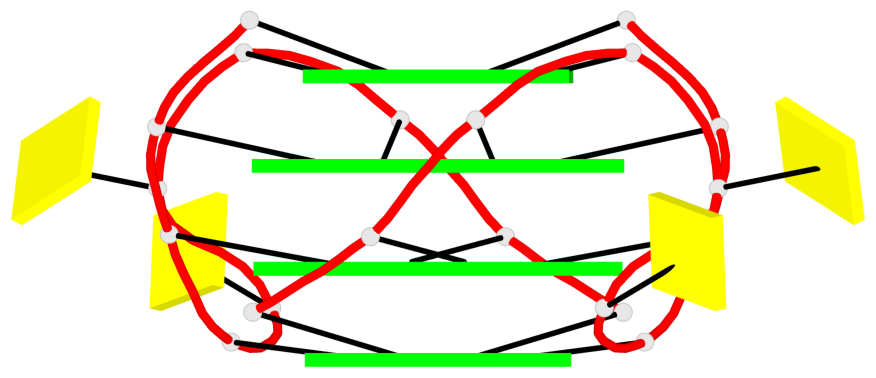
Base-block schematics in six views
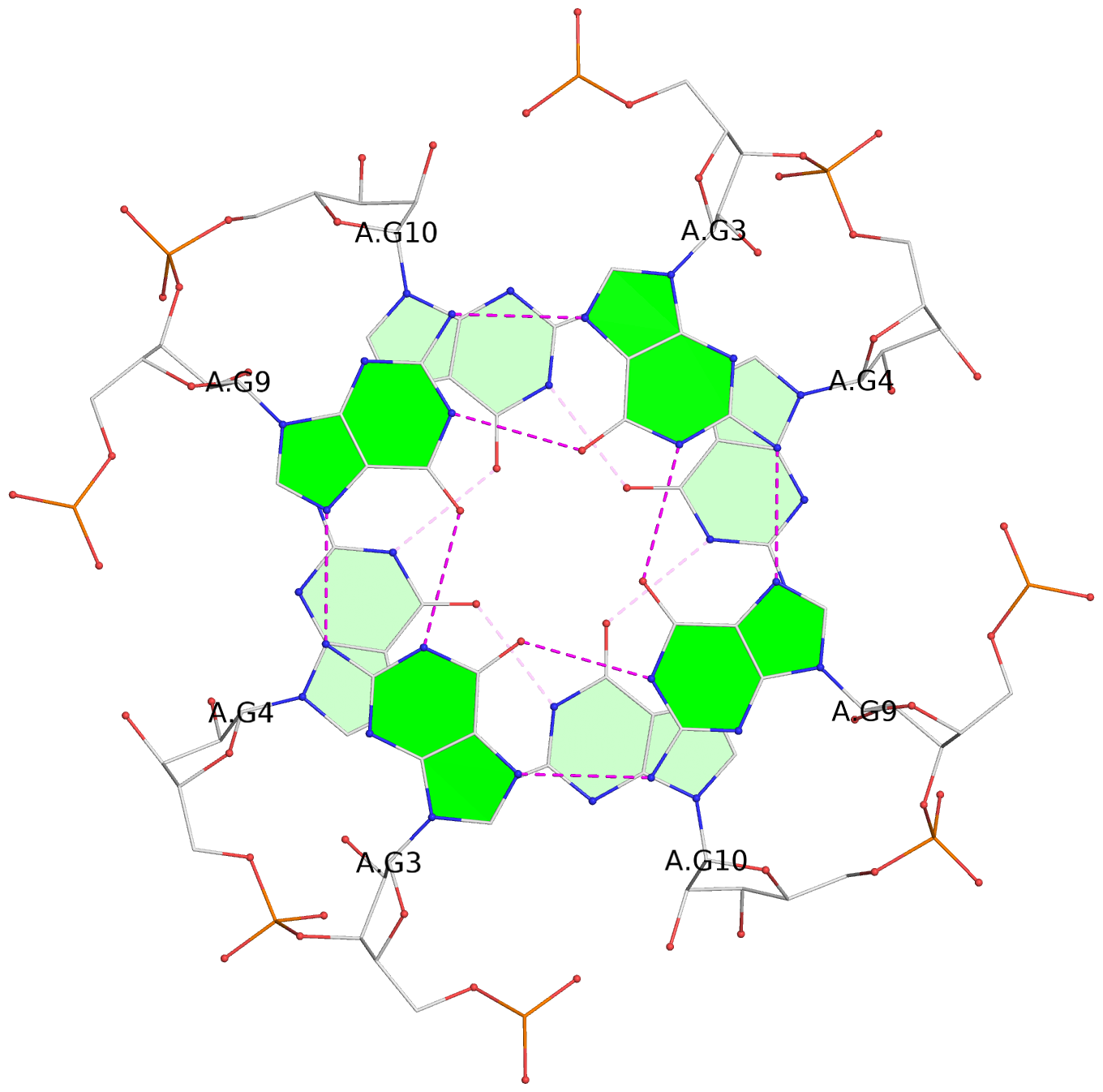
List of 8 G-tetrads
1 glyco-bond=---- sugar=3-3- groove=---- planarity=0.151 type=planar nts=4 GGGG 1:A.G1,1:A.G7,2:A.G1,2:A.G7 2 glyco-bond=---- sugar=3333 groove=---- planarity=0.167 type=other nts=4 GGGG 1:A.G2,1:A.G8,2:A.G2,2:A.G8 3 glyco-bond=---- sugar=---- groove=---- planarity=0.161 type=other nts=4 GGGG 1:A.G3,1:A.G9,2:A.G3,2:A.G9 4 glyco-bond=---- sugar=-3-3 groove=---- planarity=0.101 type=planar nts=4 GGGG 1:A.G4,1:A.G10,2:A.G4,2:A.G10 5 glyco-bond=---- sugar=3-3- groove=---- planarity=0.164 type=other nts=4 GGGG 1:B.G1,1:B.G7,2:B.G1,2:B.G7 6 glyco-bond=---- sugar=3333 groove=---- planarity=0.187 type=other nts=4 GGGG 1:B.G2,1:B.G8,2:B.G2,2:B.G8 7 glyco-bond=---- sugar=---- groove=---- planarity=0.134 type=planar nts=4 GGGG 1:B.G3,1:B.G9,2:B.G3,2:B.G9 8 glyco-bond=---- sugar=-3-3 groove=---- planarity=0.070 type=planar nts=4 GGGG 1:B.G4,1:B.G10,2:B.G4,2:B.G10
List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 8 G-tetrad layers, inter-molecular, with 2 stems
List of 2 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 4 G-tetrad layers, 2 loops, inter-molecular, UUUU, parallel, parallel(4+0)
Stem#2, 4 G-tetrad layers, 2 loops, inter-molecular, UUUU, parallel, parallel(4+0)
List of 1 G4 coaxial stack
1 G4 helix#1 contains 2 G4 stems: [#1,#2] [5'/5']