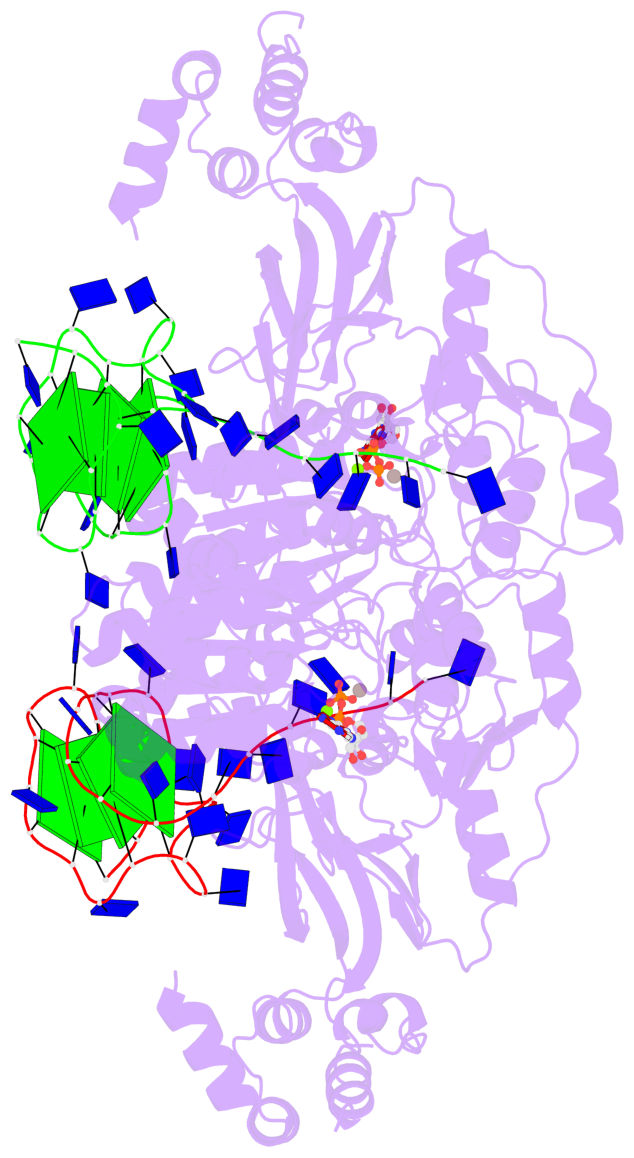
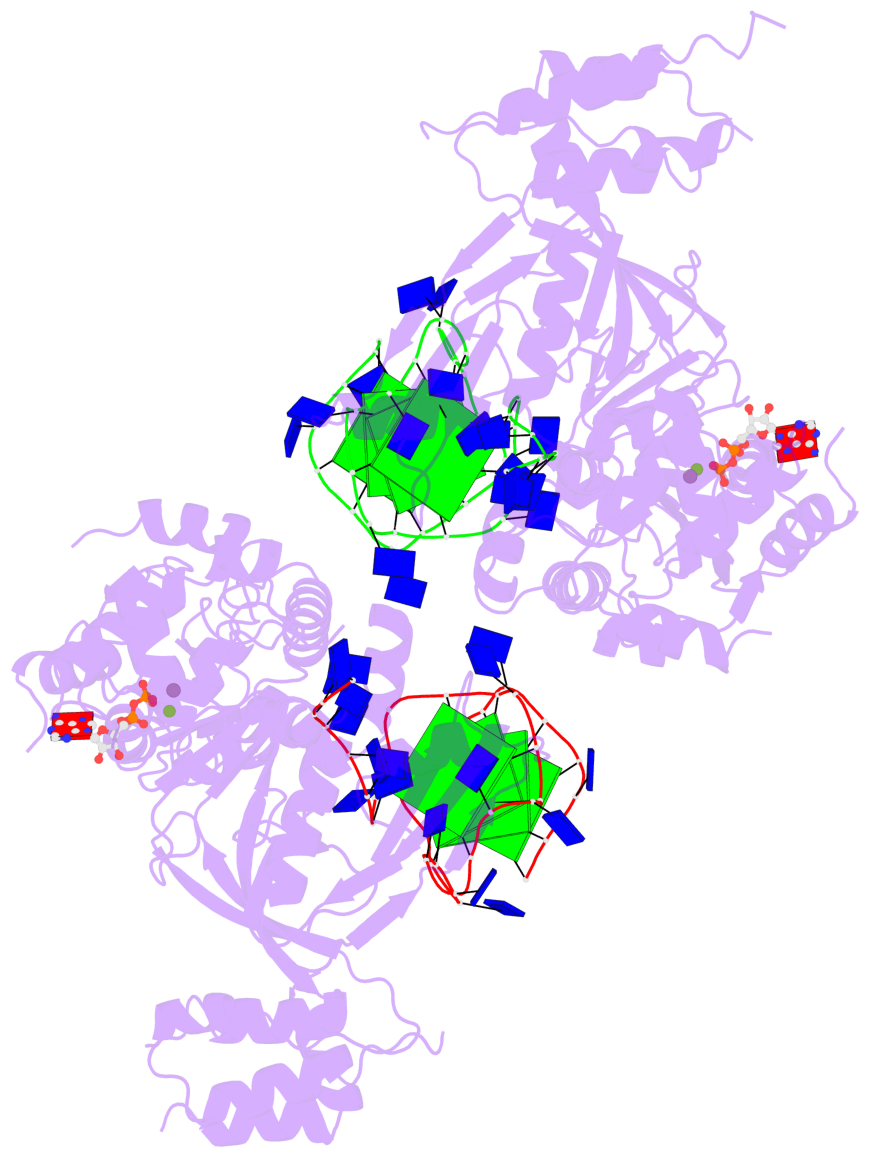
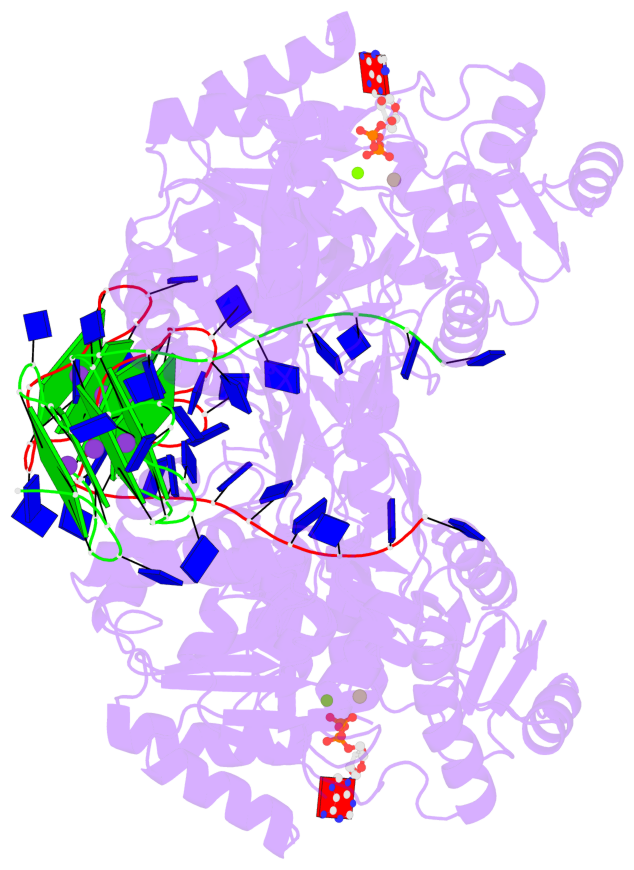
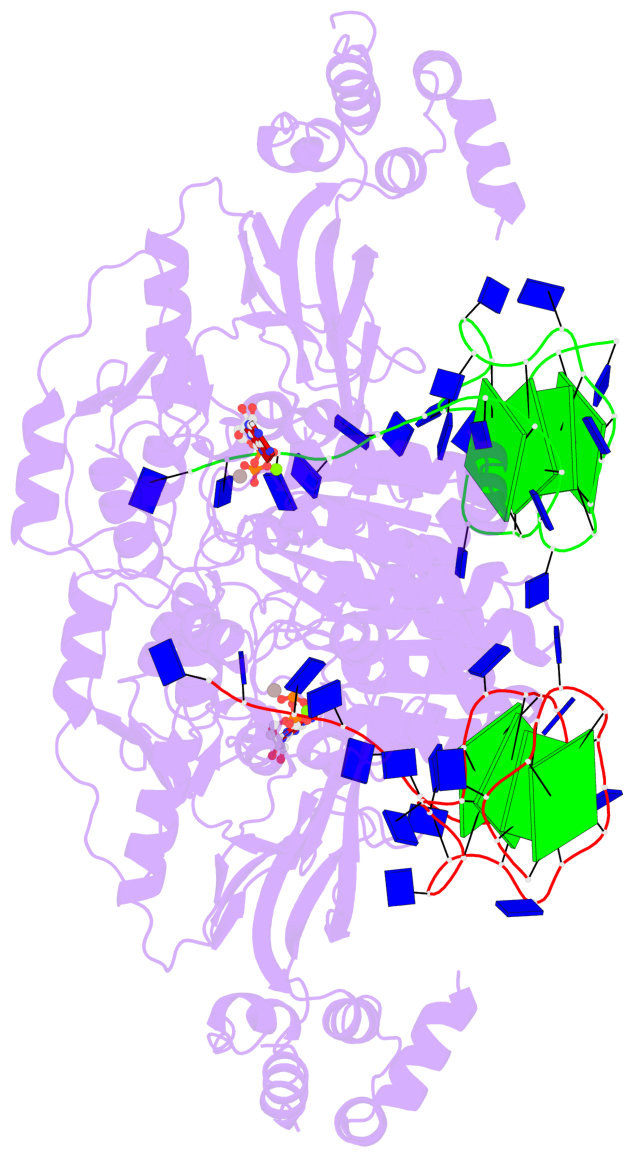
Detailed DSSR results for the G-quadruplex: PDB entry 8xak
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 8xak
- Class
- hydrolase-DNA
- Method
- X-ray (3.5 Å)
- Summary
- Structure of pif1-g4 complex
- Reference
- Hong Z, Byrd AK, Gao J, Das P, Tan VQ, Malone EG, Osei B, Marecki JC, Protacio RU, Wahls WP, Raney KD, Song H (2024): "Eukaryotic Pif1 helicase unwinds G-quadruplex and dsDNA using a conserved wedge." Nat Commun, 15, 6104. doi: 10.1038/s41467-024-50575-8.
- Abstract
- G-quadruplexes (G4s) formed by guanine-rich nucleic acids induce genome instability through impeding DNA replication fork progression. G4s are stable DNA structures, the unfolding of which require the functions of DNA helicases. Pif1 helicase binds preferentially to G4 DNA and plays multiple roles in maintaining genome stability, but the mechanism by which Pif1 unfolds G4s is poorly understood. Here we report the co-crystal structure of Saccharomyces cerevisiae Pif1 (ScPif1) bound to a G4 DNA with a 5' single-stranded DNA (ssDNA) segment. Unlike the Thermus oshimai Pif1-G4 structure, in which the 1B and 2B domains confer G4 recognition, ScPif1 recognizes G4 mainly through the wedge region in the 1A domain that contacts the 5' most G-tetrad directly. A conserved Arg residue in the wedge is required for Okazaki fragment processing but not for mitochondrial function or for suppression of gross chromosomal rearrangements. Multiple substitutions at this position have similar effects on resolution of DNA duplexes and G4s, suggesting that ScPif1 may use the same wedge to unwind G4 and dsDNA. Our results reveal the mechanism governing dsDNA unwinding and G4 unfolding by ScPif1 helicase that can potentially be generalized to other eukaryotic Pif1 helicases and beyond.
- G4 notes
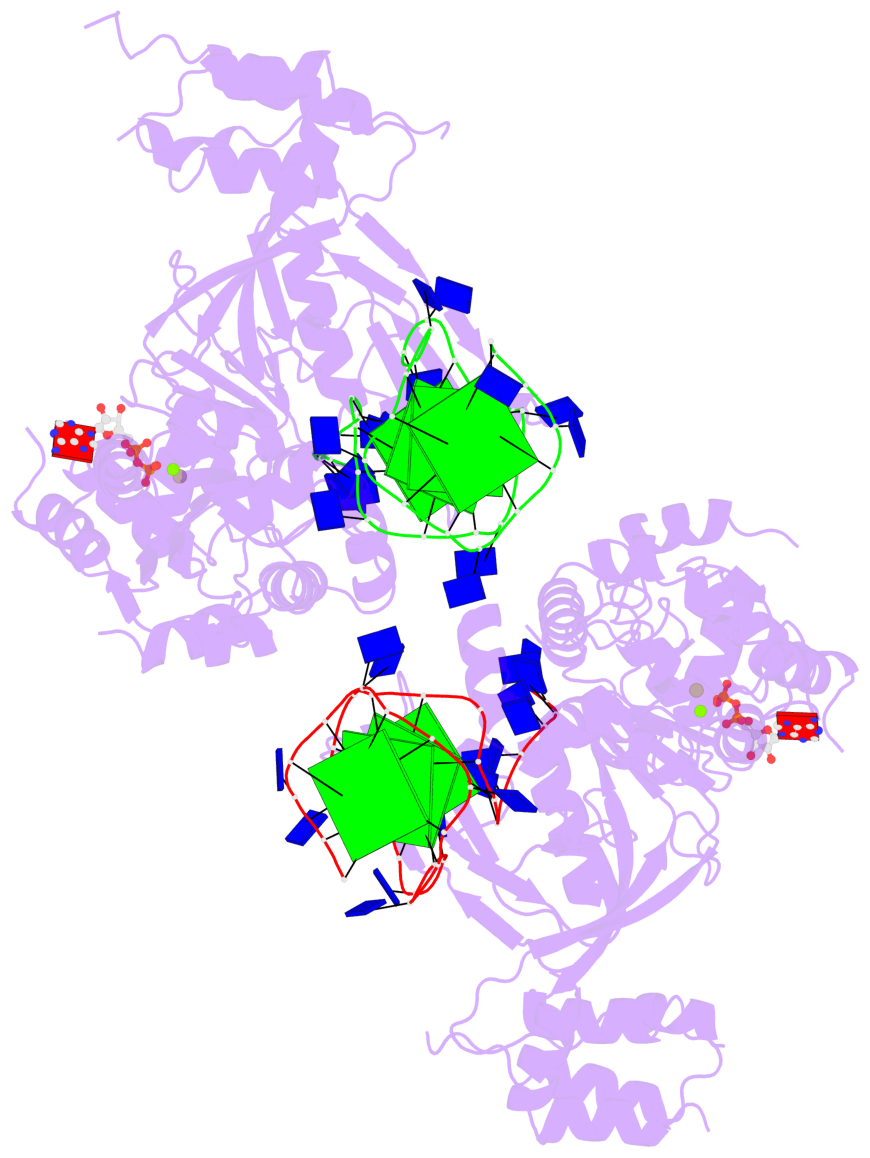
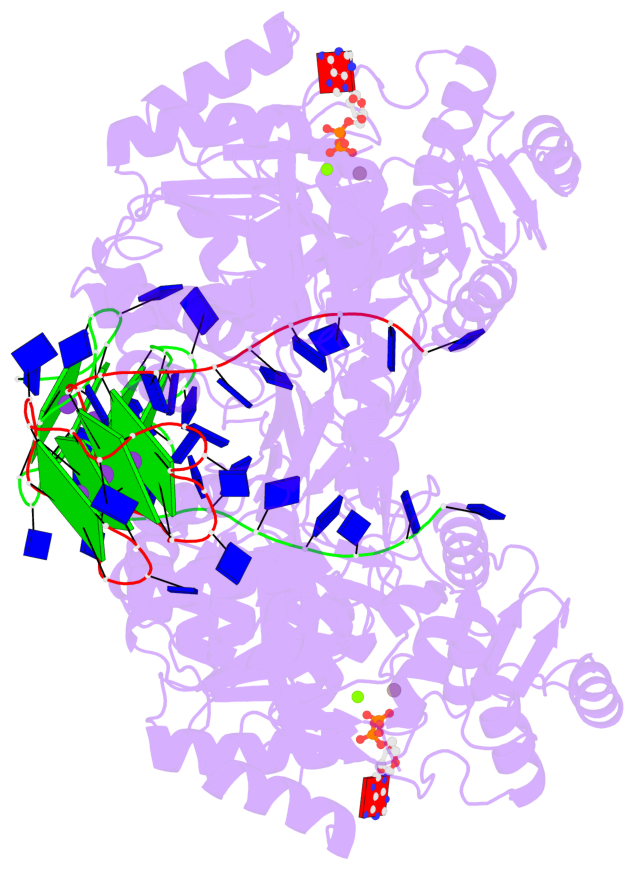
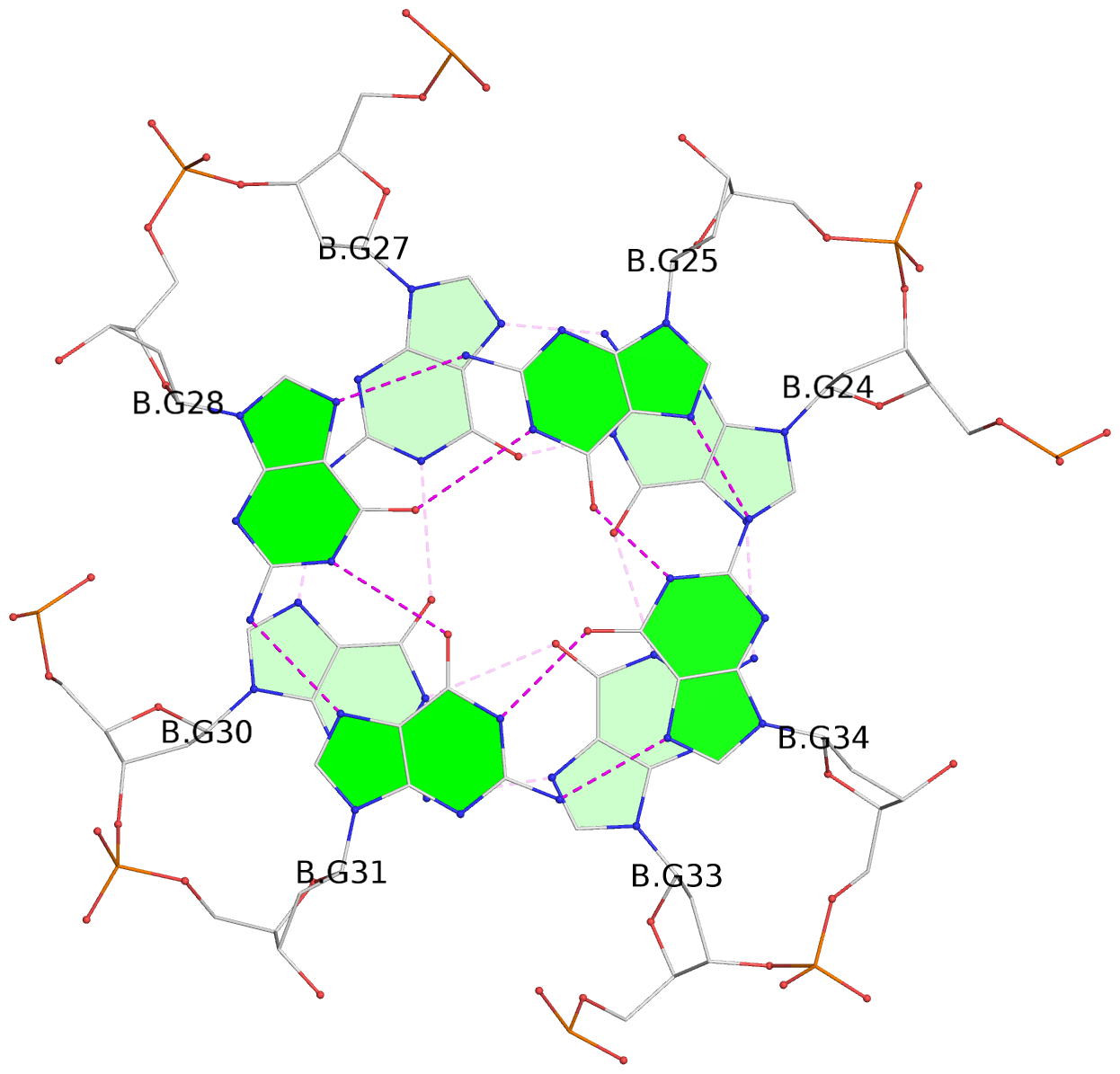
- 8 G-tetrads, 2 G4 helices, 4 G4 stems, 2 G4 coaxial stacks, 2(-P-P-P), parallel(4+0), UUUU, coaxial interfaces: 3'/5'
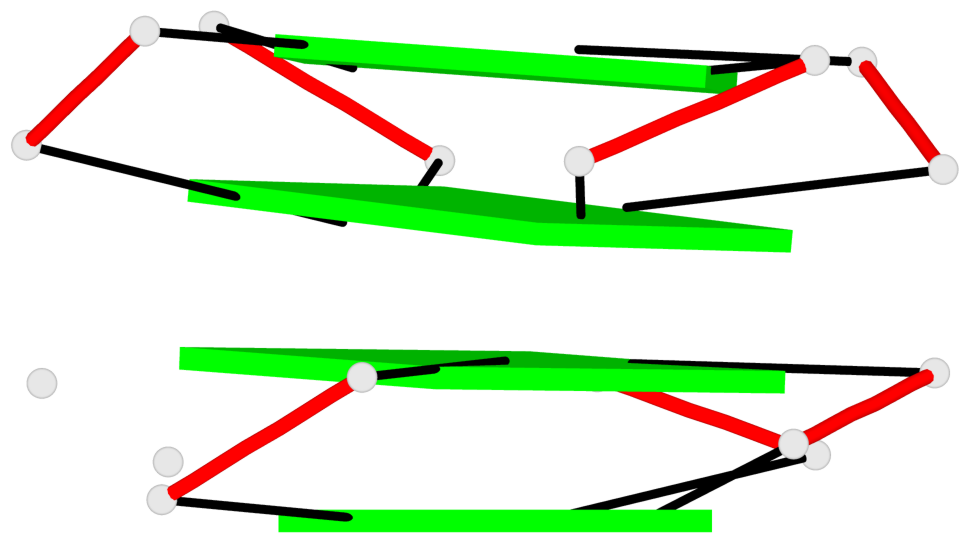
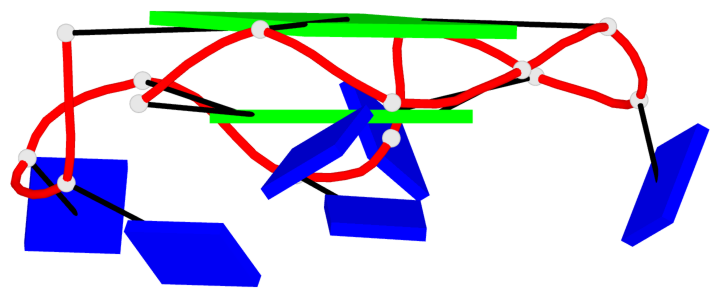
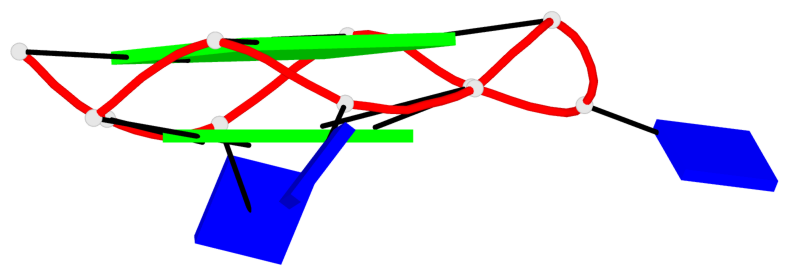
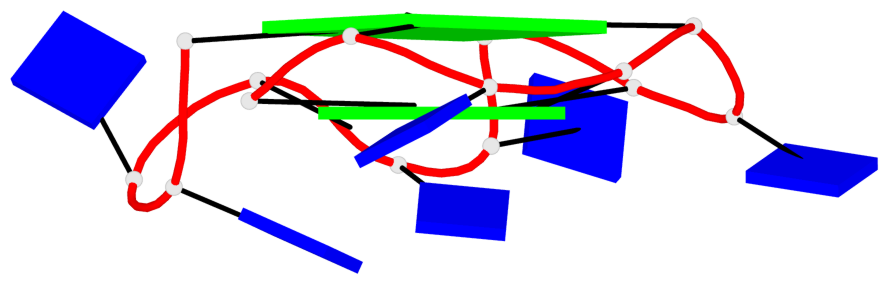
Base-block schematics in six views
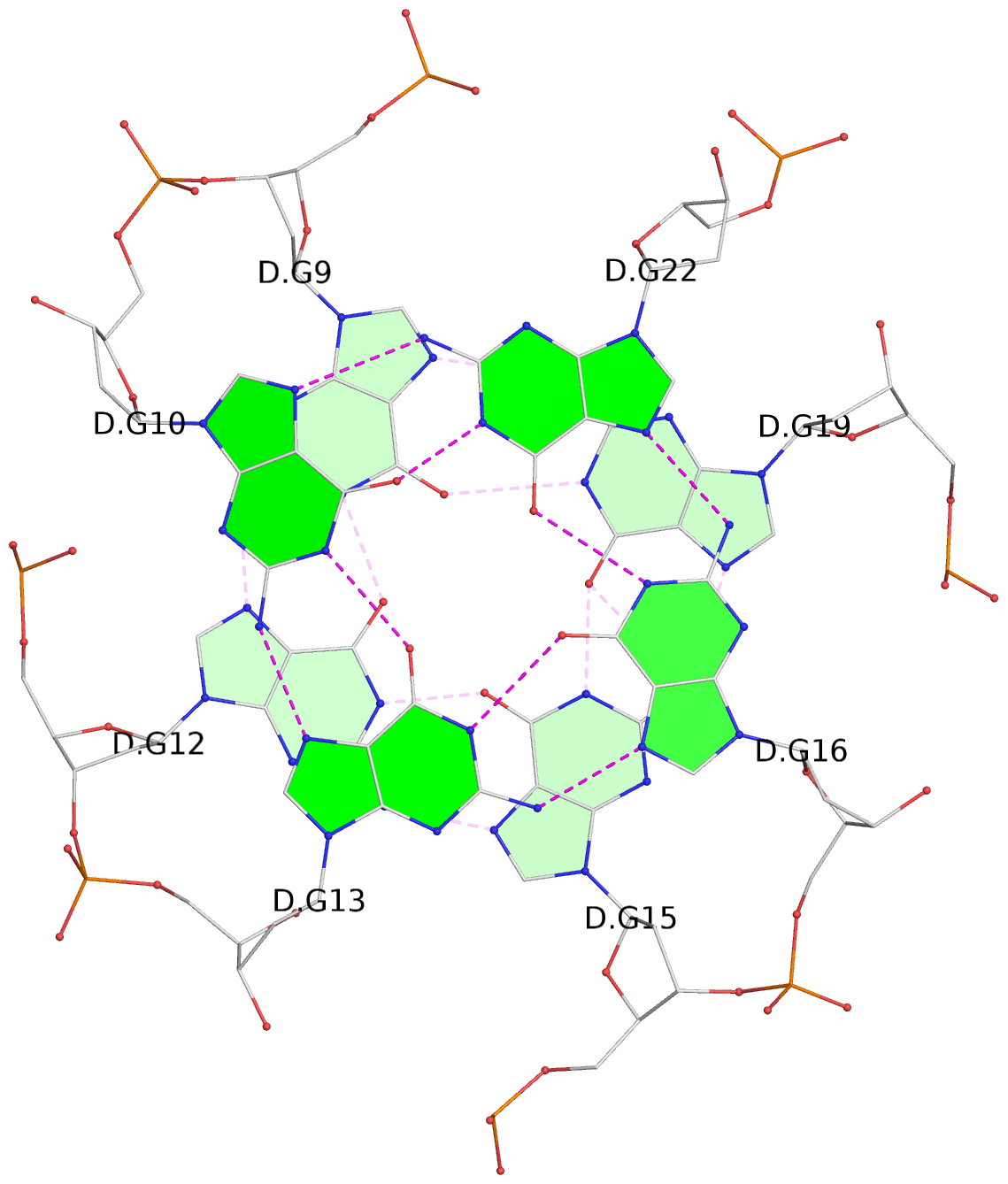
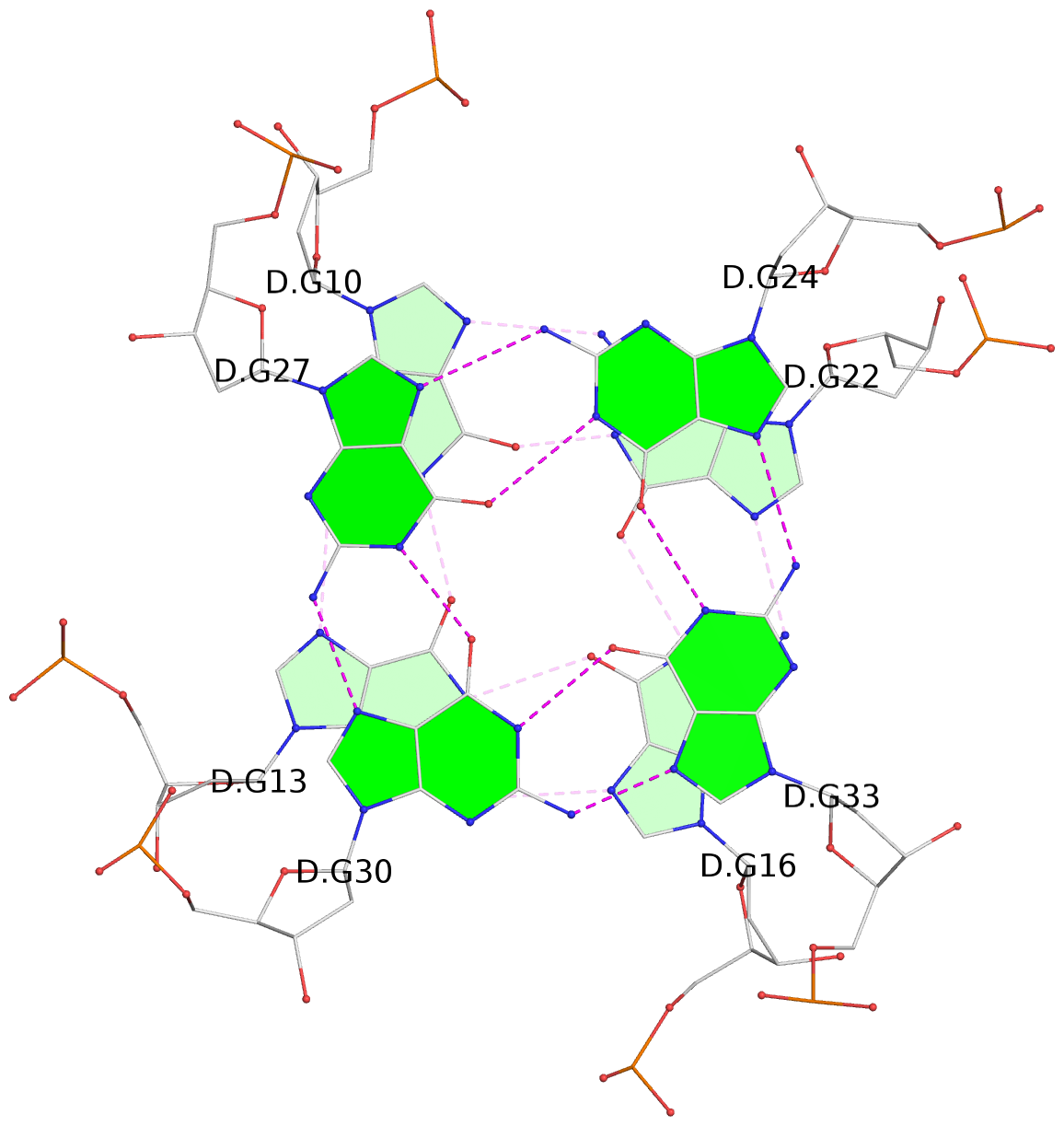
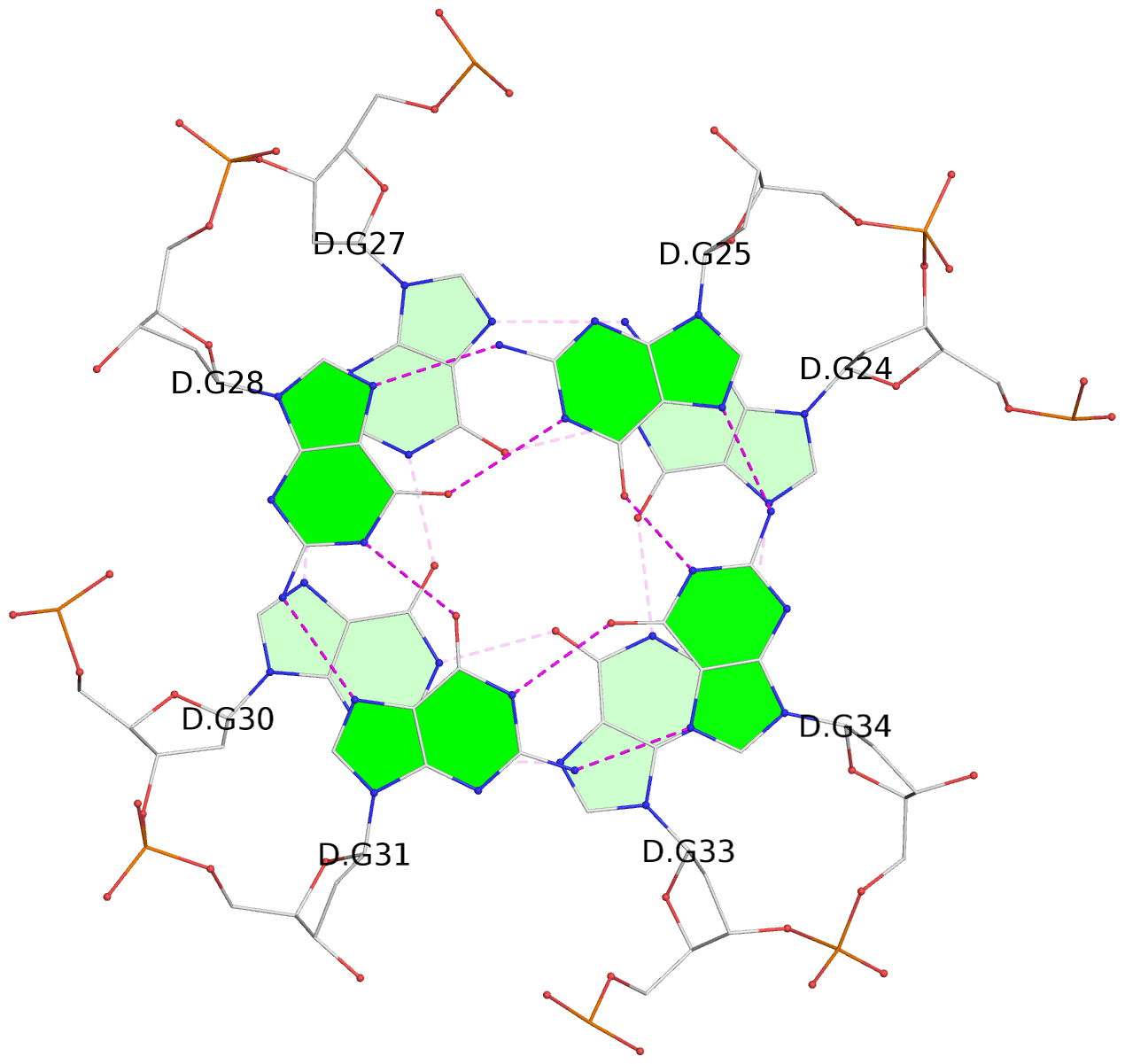
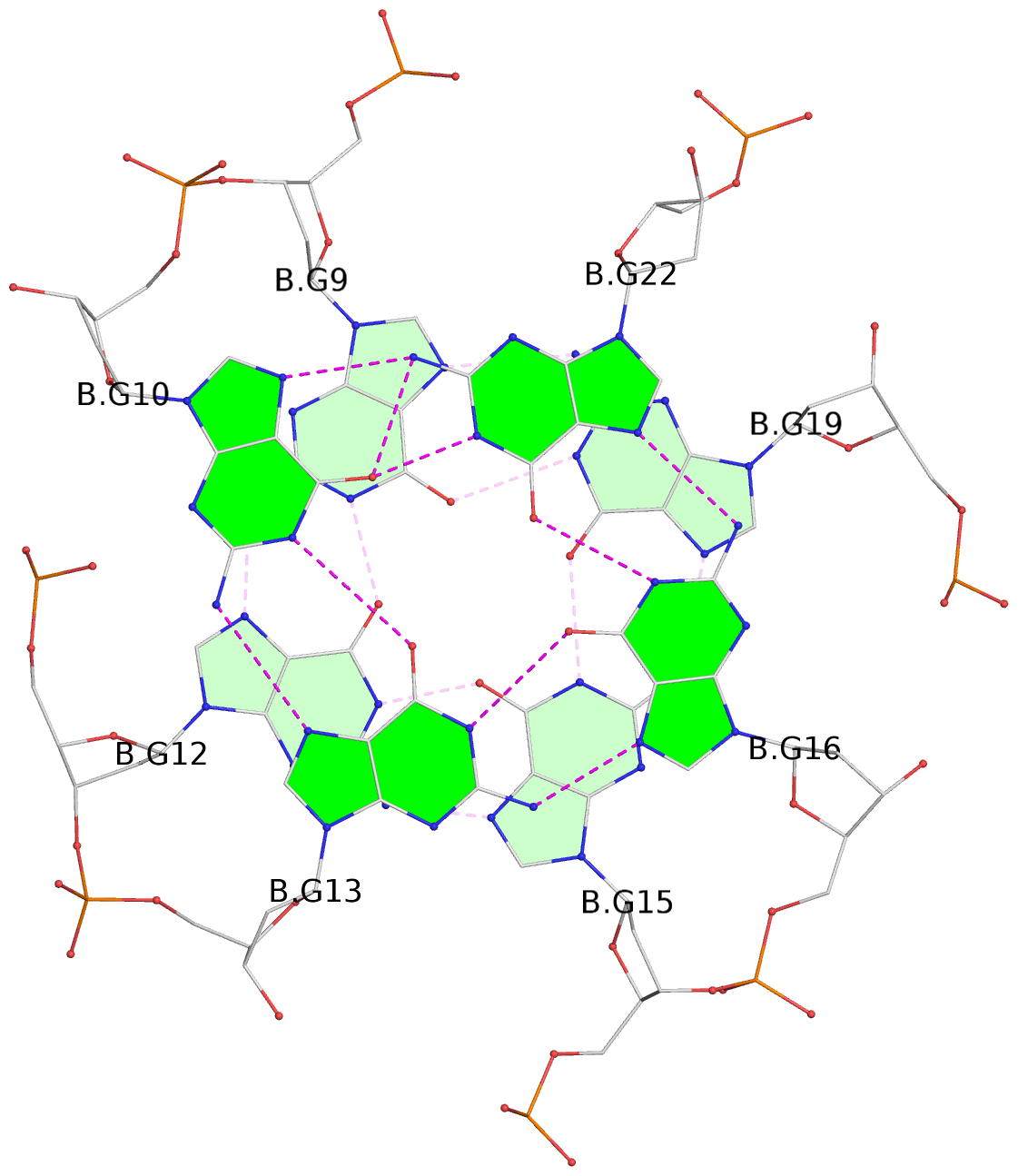
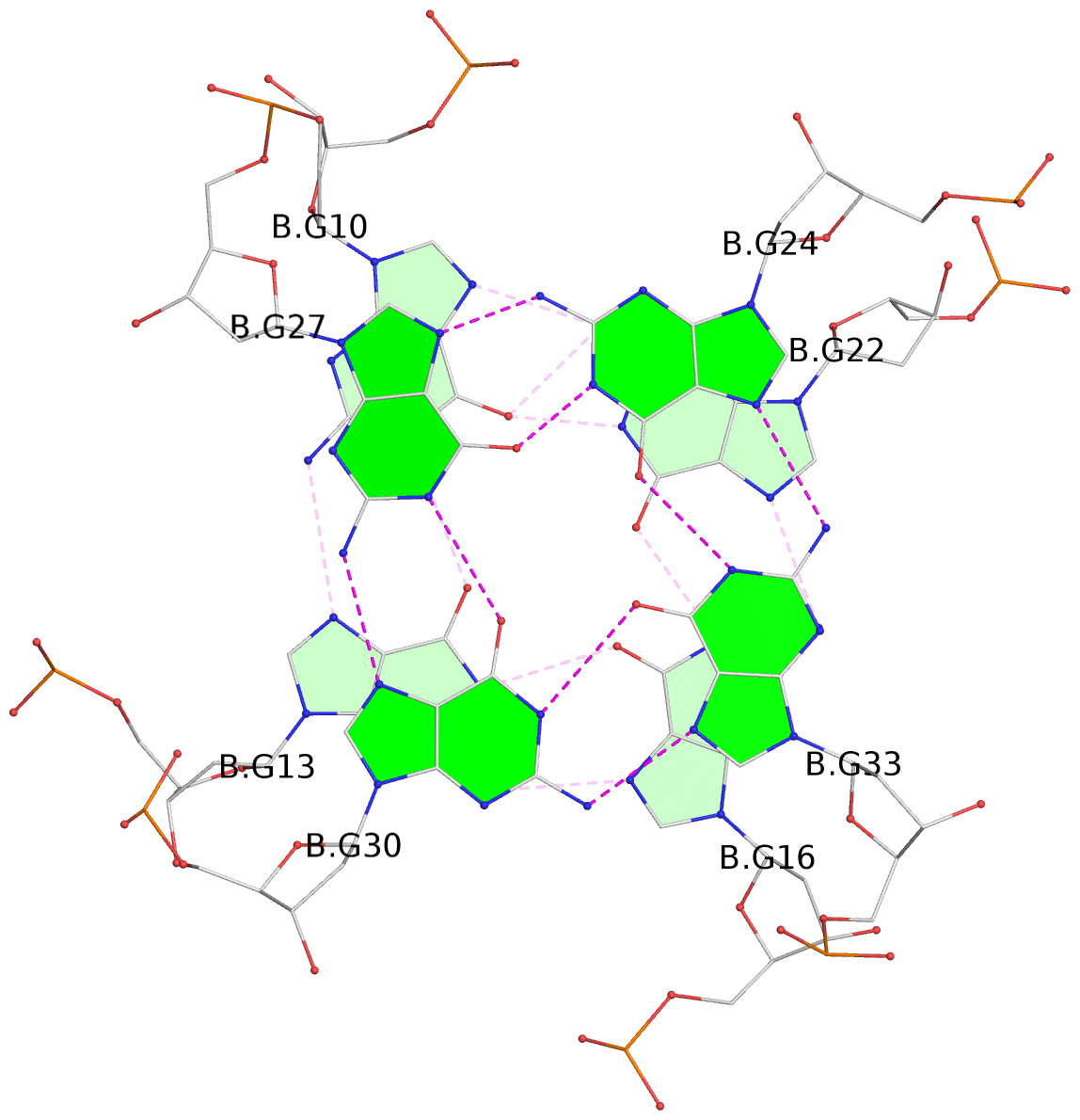
List of 8 G-tetrads
1 glyco-bond=---- sugar=---- groove=---- planarity=0.168 type=other nts=4 GGGG D.DG9,D.DG12,D.DG15,D.DG19 2 glyco-bond=---- sugar=3--- groove=---- planarity=0.231 type=other nts=4 GGGG D.DG10,D.DG13,D.DG16,D.DG22 3 glyco-bond=---- sugar=-..- groove=---- planarity=0.213 type=other nts=4 GGGG D.DG24,D.DG27,D.DG30,D.DG33 4 glyco-bond=---- sugar=---. groove=---- planarity=0.229 type=other nts=4 GGGG D.DG25,D.DG28,D.DG31,D.DG34 5 glyco-bond=---- sugar=---- groove=---- planarity=0.204 type=other nts=4 GGGG B.DG9,B.DG12,B.DG15,B.DG19 6 glyco-bond=---- sugar=.--- groove=---- planarity=0.263 type=other nts=4 GGGG B.DG10,B.DG13,B.DG16,B.DG22 7 glyco-bond=---- sugar=---- groove=---- planarity=0.208 type=other nts=4 GGGG B.DG24,B.DG27,B.DG30,B.DG33 8 glyco-bond=---- sugar=---- groove=---- planarity=0.218 type=other nts=4 GGGG B.DG25,B.DG28,B.DG31,B.DG34
List of 2 G4-helices
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 4 G-tetrad layers, INTRA-molecular, with 2 stems
Helix#2, 4 G-tetrad layers, INTRA-molecular, with 2 stems
List of 4 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(-P-P-P), parallel(4+0)
Stem#2, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(-P-P-P), parallel(4+0)
Stem#3, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(-P-P-P), parallel(4+0)
Stem#4, 2 G-tetrad layers, 3 loops, INTRA-molecular, UUUU, parallel, 2(-P-P-P), parallel(4+0)
List of 2 G4 coaxial stacks
1 G4 helix#1 contains 2 G4 stems: [#1,#2] [3'/5'] 2 G4 helix#2 contains 2 G4 stems: [#3,#4] [3'/5']