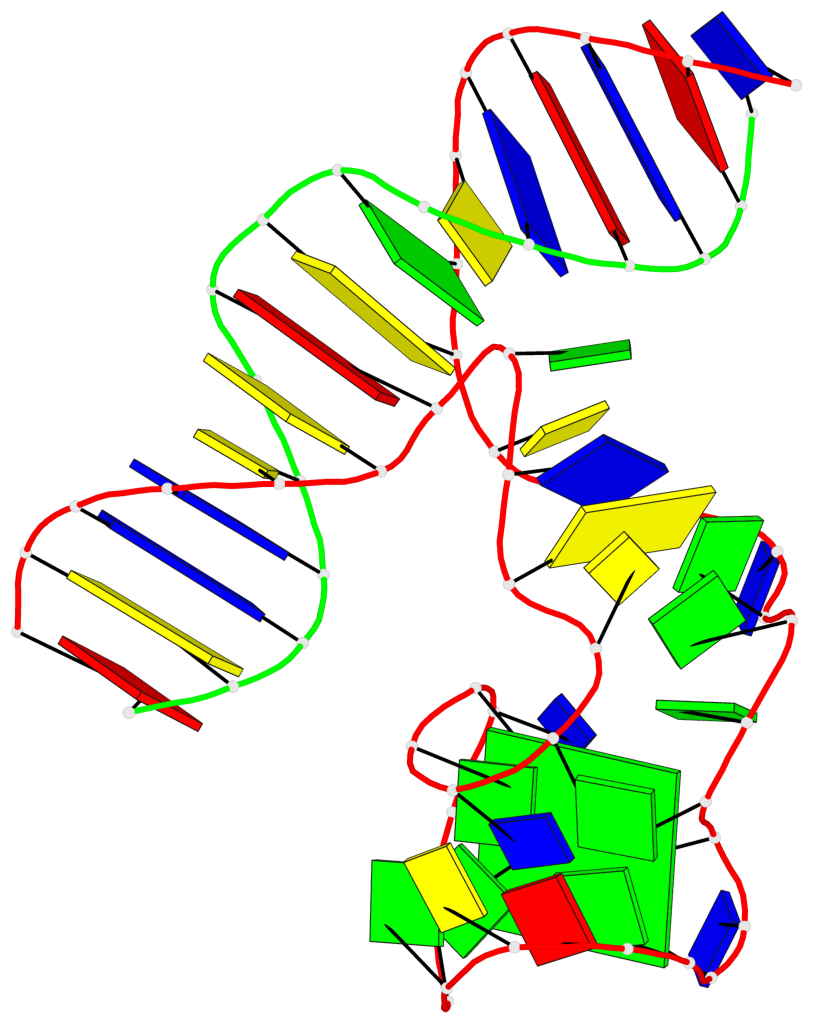
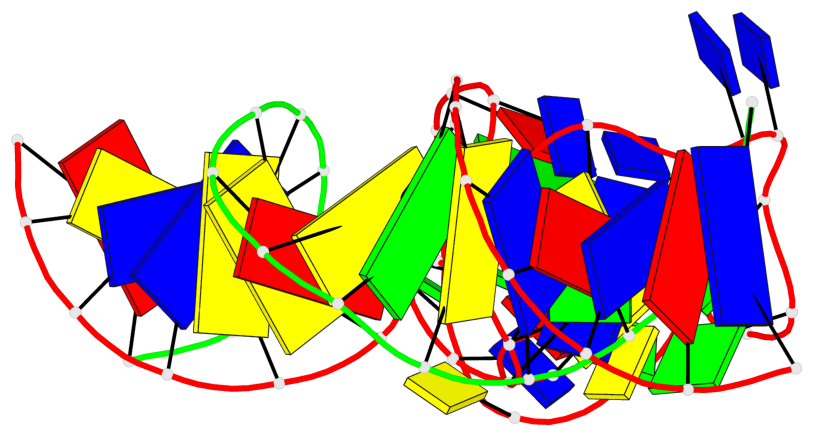
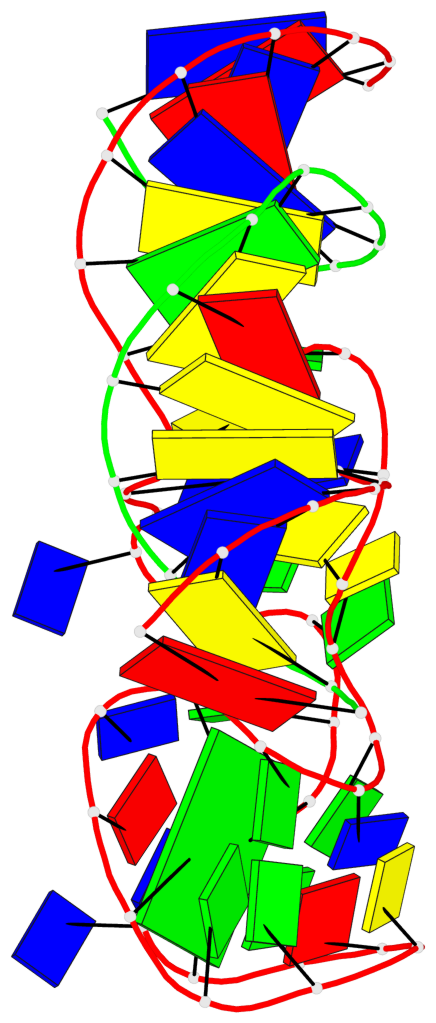
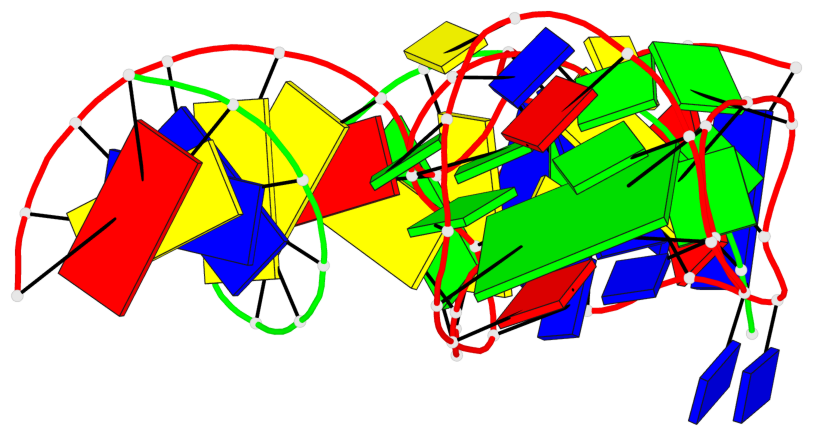
Detailed DSSR results for the G-quadruplex: PDB entry 9eoq
Created and maintained by Xiang-Jun Lu <xiangjun@x3dna.org>
Citation: Please cite the NAR'20 DSSR-PyMOL schematics paper and/or the NAR'15 DSSR method paper.
Summary information
- PDB id
- 9eoq
- Class
- DNA
- Method
- cryo-EM (7.5 Å)
- Summary
- cryo-EM structure of a 1033 scaffold base DNA origami nanostructure v4 and tba
- Reference
- Khoshouei A, Kempf G, Mykhailiuk V, Griessing JM, Honemann MN, Kater L, Cavadini S, Dietz H (2024): "Designing Rigid DNA Origami Templates for Molecular Visualization Using Cryo-EM." Nano Lett., 24, 5031-5038. doi: 10.1021/acs.nanolett.4c00915.
- Abstract
- DNA origami, a method for constructing nanostructures from DNA, offers potential for diverse scientific and technological applications due to its ability to integrate various molecular functionalities in a programmable manner. In this study, we examined the impact of internal crossover distribution and the compositional uniformity of staple strands on the structure of multilayer DNA origami using cryogenic electron microscopy (cryo-EM) single-particle analysis. A refined DNA object was utilized as an alignment framework in a host-guest model, where we successfully resolved an 8 kDa thrombin binding aptamer (TBA) linked to the host object. Our results broaden the spectrum of DNA in structural applications.
- G4 notes
- 1 G-tetrad
Base-block schematics in six views
List of 1 G-tetrad
1 glyco-bond=-s-s sugar=-..- groove=wnwn planarity=0.389 type=bowl-2 nts=4 GGGG A.DG16,A.DG28,A.DG25,A.DG19
List of 3 non-stem G4-loops (including the two closing Gs)
1 type=lateral helix=#-1 nts=4 GTAG A.DG16,A.DT17,A.DA18,A.DG19 2 type=lateral helix=#-1 nts=7 GGGCAGG A.DG19,A.DG20,A.DG21,A.DC22,A.DA23,A.DG24,A.DG25 3 type=lateral helix=#-1 nts=4 GTTG A.DG25,A.DT26,A.DT27,A.DG28