DSSR-enabled G4 identification/annotation: PDB entry 8k85
Summary information and primary citation
- PDB-id
- 8k85
- Class
- RNA
- Method
- X-ray (2.95 Å)
- Summary
- Crystal structure of red broccoli aptamer with obi
- Reference
- Peng X, Huang L: "Structural analysis of various Broccoli aptamers with different Fluorophores."
- Abstract
- G4 notes
- 4 G-tetrads, 2 G4 helices, 2 G4 stems, UUDD, anti-parallel:basket2, 2+2
Cartoon-block schematics in six views (download the tarball)
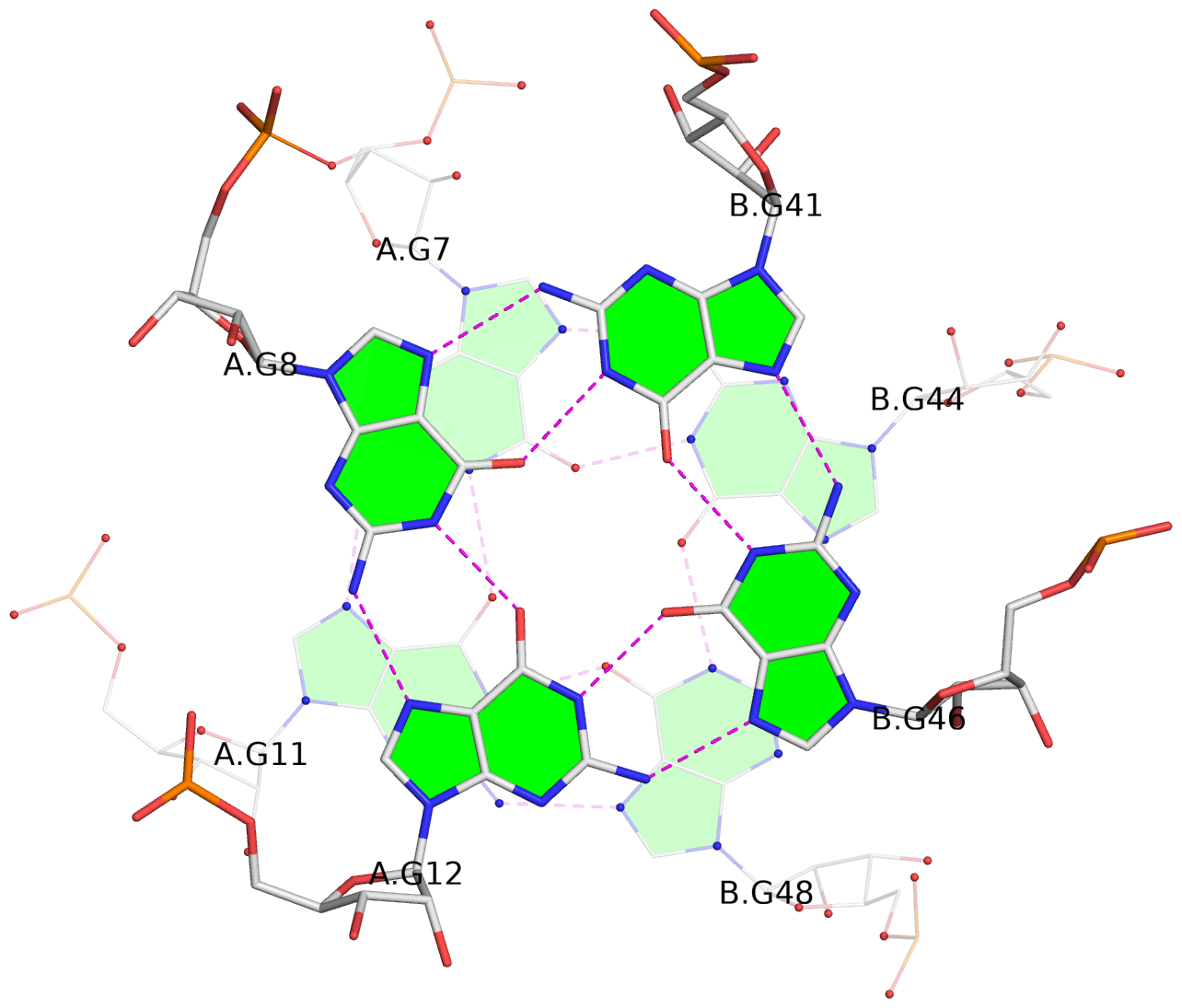
List of 4 G-tetrads
1 glyco-bond=--s- sugar=-33. groove=-wn- planarity=0.290 type=other -- nts=4 GGGG 1:A.G7,1:A.G11,1:B.G48,1:B.G44 2 glyco-bond=--ss sugar=-3-3 groove=-w-n planarity=0.155 type=planar -- nts=4 GGGG 1:A.G8,1:A.G12,1:B.G46,1:B.G41 3 glyco-bond=ss-- sugar=3-3- groove=-w-n planarity=0.176 type=other -- nts=4 GGGG 1:A.G41,1:A.G46,1:B.G12,1:B.G8 4 glyco-bond=---s sugar=.-33 groove=--wn planarity=0.330 type=other -- nts=4 GGGG 1:A.G44,1:B.G7,1:B.G11,1:A.G48
List of 2 G4-helices
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 2 G-tetrads, inter-molecular, with 1 stem
Helix#2, 2 G-tetrads, inter-molecular, with 1 stem
List of 2 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.