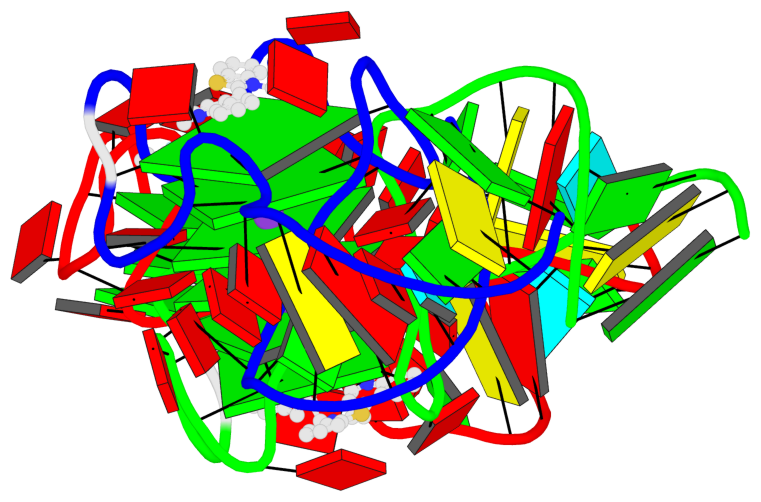
DSSR-enabled G4 identification/annotation: PDB entry 8vxx
Summary information and primary citation
- PDB-id
- 8vxx
- Class
- RNA
- Method
- X-ray (3.1 Å)
- Summary
- Mango ii bound to 365a-061
- Reference
- Passalacqua LFM, Ferre-D'Amare AR, Schneekloth JS: "Mango II bound to 365A-061."
- Abstract
- G4 notes
- 9 G-tetrads, 2 G4 helices, 3 G4 stems, 1 G4 coaxial stack, UUUU, parallel, 4+0, 3(-P-P-P), coaxial interfaces: 5'/5'
Cartoon-block schematics in six views (download the tarball)
List of 9 G-tetrads
1 glyco-bond=---- sugar=-3-3 groove=---- planarity=0.263 type=other O+ nts=4 GGGG 1:A.G10,1:A.G15,1:A.G20,1:A.G26 2 glyco-bond=---- sugar=3333 groove=---- planarity=0.166 type=other O+ nts=4 GGGG 1:A.G11,1:A.G16,1:A.G21,1:A.G27 3 glyco-bond=---- sugar=---- groove=---- planarity=0.122 type=planar O- nts=4 GGGG 1:A.G13,1:A.G29,1:A.G24,1:A.G18 4 glyco-bond=---- sugar=-3-3 groove=---- planarity=0.345 type=other O+ nts=4 GGGG 1:B.G10,1:B.G15,1:B.G20,1:B.G26 5 glyco-bond=---- sugar=3333 groove=---- planarity=0.156 type=planar O+ nts=4 GGGG 1:B.G11,1:B.G16,1:B.G21,1:B.G27 6 glyco-bond=---- sugar=---- groove=---- planarity=0.176 type=other O- nts=4 GGGG 1:B.G13,1:B.G29,1:B.G24,1:B.G18 7 glyco-bond=---- sugar=33-3 groove=---- planarity=0.289 type=other O+ nts=4 GGGG 1:C.G10,1:C.G15,1:C.G20,1:C.G26 8 glyco-bond=---- sugar=3333 groove=---- planarity=0.101 type=planar O+ nts=4 GGGG 1:C.G11,1:C.G16,1:C.G21,1:C.G27 9 glyco-bond=---- sugar=---- groove=---- planarity=0.181 type=other O- nts=4 GGGG 1:C.G13,1:C.G29,1:C.G24,1:C.G18
List of 2 G4-helices
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem.
Helix#1, 3 G-tetrads, INTRA-molecular, with 1 stem
Helix#2, 6 G-tetrads, inter-molecular, with 2 stems
List of 3 G4-stems
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands.
Stem#1, 3 G-tetrads, 3 loops, INTRA-molecular, UUUU, parallel, 4+0, 3(-P-P-P)
Stem#2, 3 G-tetrads, 3 loops, INTRA-molecular, UUUU, parallel, 4+0, 3(-P-P-P)
Stem#3, 3 G-tetrads, 3 loops, INTRA-molecular, UUUU, parallel, 4+0, 3(-P-P-P)
List of 1 G4 coaxial stack
1 G4 helix#2 contains 2 G4 stems: [#2,#3] [5'/5']